MitoRes: a resource of nuclear-encoded mitochondrial genes and their products in Metazoa - PubMed (original) (raw)
Comparative Study
MitoRes: a resource of nuclear-encoded mitochondrial genes and their products in Metazoa
Domenico Catalano et al. BMC Bioinformatics. 2006.
Abstract
Background: Mitochondria are sub-cellular organelles that have a central role in energy production and in other metabolic pathways of all eukaryotic respiring cells. In the last few years, with more and more genomes being sequenced, a huge amount of data has been generated providing an unprecedented opportunity to use the comparative analysis approach in studies of evolution and functional genomics with the aim of shedding light on molecular mechanisms regulating mitochondrial biogenesis and metabolism. In this context, the problem of the optimal extraction of representative datasets of genomic and proteomic data assumes a crucial importance. Specialised resources for nuclear-encoded mitochondria-related proteins already exist; however, no mitochondrial database is currently available with the same features of MitoRes, which is an update of the MitoNuc database extensively modified in its structure, data sources and graphical interface. It contains data on nuclear-encoded mitochondria-related products for any metazoan species for which this type of data is available and also provides comprehensive sequence datasets (gene, transcript and protein) as well as useful tools for their extraction and export.
Description: MitoRes http://www2.ba.itb.cnr.it/MitoRes/ consolidates information from publicly external sources and automatically annotates them into a relational database. Additionally, it also clusters proteins on the basis of their sequence similarity and interconnects them with genomic data. The search engine and sequence management tools allow the query/retrieval of the database content and the extraction and export of sequences (gene, transcript, protein) and related sub-sequences (intron, exon, UTR, CDS, signal peptide and gene flanking regions) ready to be used for in silico analysis.
Conclusion: The tool we describe here has been developed to support lab scientists and bioinformaticians alike in the characterization of molecular features and evolution of mitochondrial targeting sequences. The way it provides for the retrieval and extraction of sequences allows the user to overcome the obstacles encountered in the integrative use of different bioinformatic resources and the completeness of the sequence collection allows intra- and interspecies comparison at different biological levels (gene, transcript and protein).
Figures
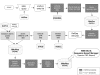
Figure 1
Flowchart depicting the process used to generate MitoRes. Dark and clear grey boxes are for BioPerl and C/C++ procedures respectively.
Figure 2
Database search form, query result page and sequence export form. A) Search form. The "i" button provides information on how the search field can be used. The "Show" check box provides the possibility to include (checked) or exclude (unchecked) information on the relevant field in the query result page. The search fields "Entry name", "Accession Number", "Species", "Chromosome", "Gene name" and database "Cross-referencing" accept lists of search terms. B) Query result page. Results are displayed as a table summarising information on retrieved entry on the basis of data field chosen in the query form. The "View" button, when pressed, shows the MitoRes entry view. The "Select page"/"Deselect page" buttons allows the selection/deselection of retrieved records for the export of associated sequences. The "Export selected" and "Export all" buttons provide access to the sequence export form for the extraction and export of selected records or of all retrieved records respectively. C) Sequence export form. Selected record/s for sequence export is/are listed at the top. The "i" button provides information on how the extraction of each sequence can be performed. Check boxes at the bottom allow users to choose the mode and sequence file format for export.
Figure 3
Cluster entry view. The Clusters list is accessible clicking the "Cluster" button in the MitoRes home page and/or the "Associated Cluster" button in the MitoRes entries. The "Export all" button at the top, allows the access to the sequence export form to perform the extraction and export of sequences associated to all the Cluster members.
Similar articles
- The MitoDrome database annotates and compares the OXPHOS nuclear genes of Drosophila melanogaster, Drosophila pseudoobscura and Anopheles gambiae.
D'Elia D, Catalano D, Licciulli F, Turi A, Tripoli G, Porcelli D, Saccone C, Caggese C. D'Elia D, et al. Mitochondrion. 2006 Oct;6(5):252-7. doi: 10.1016/j.mito.2006.07.001. Epub 2006 Jul 21. Mitochondrion. 2006. PMID: 16982216 - PCHM: A bioinformatic resource for high-throughput human mitochondrial proteome searching and comparison.
Kim T, Kim E, Park SJ, Joo H. Kim T, et al. Comput Biol Med. 2009 Aug;39(8):689-96. doi: 10.1016/j.compbiomed.2009.05.006. Epub 2009 Jun 21. Comput Biol Med. 2009. PMID: 19541297 - Genome annotation of Anopheles gambiae using mass spectrometry-derived data.
Kalume DE, Peri S, Reddy R, Zhong J, Okulate M, Kumar N, Pandey A. Kalume DE, et al. BMC Genomics. 2005 Sep 19;6:128. doi: 10.1186/1471-2164-6-128. BMC Genomics. 2005. PMID: 16171517 Free PMC article. - Advances in the Exon-Intron Database (EID).
Shepelev V, Fedorov A. Shepelev V, et al. Brief Bioinform. 2006 Jun;7(2):178-85. doi: 10.1093/bib/bbl003. Epub 2006 Mar 9. Brief Bioinform. 2006. PMID: 16772261 Review. - TAIR: a resource for integrated Arabidopsis data.
Garcia-Hernandez M, Berardini TZ, Chen G, Crist D, Doyle A, Huala E, Knee E, Lambrecht M, Miller N, Mueller LA, Mundodi S, Reiser L, Rhee SY, Scholl R, Tacklind J, Weems DC, Wu Y, Xu I, Yoo D, Yoon J, Zhang P. Garcia-Hernandez M, et al. Funct Integr Genomics. 2002 Nov;2(6):239-53. doi: 10.1007/s10142-002-0077-z. Epub 2002 Oct 3. Funct Integr Genomics. 2002. PMID: 12444417 Review.
Cited by
- MTSviewer: A database to visualize mitochondrial targeting sequences, cleavage sites, and mutations on protein structures.
Bayne AN, Dong J, Amiri S, Farhan SMK, Trempe JF. Bayne AN, et al. PLoS One. 2023 Apr 24;18(4):e0284541. doi: 10.1371/journal.pone.0284541. eCollection 2023. PLoS One. 2023. PMID: 37093842 Free PMC article. - mitoXplorer, a visual data mining platform to systematically analyze and visualize mitochondrial expression dynamics and mutations.
Yim A, Koti P, Bonnard A, Marchiano F, Dürrbaum M, Garcia-Perez C, Villaveces J, Gamal S, Cardone G, Perocchi F, Storchova Z, Habermann BH. Yim A, et al. Nucleic Acids Res. 2020 Jan 24;48(2):605-632. doi: 10.1093/nar/gkz1128. Nucleic Acids Res. 2020. PMID: 31799603 Free PMC article. - MitProNet: A knowledgebase and analysis platform of proteome, interactome and diseases for mammalian mitochondria.
Wang J, Yang J, Mao S, Chai X, Hu Y, Hou X, Tang Y, Bi C, Li X. Wang J, et al. PLoS One. 2014 Oct 27;9(10):e111187. doi: 10.1371/journal.pone.0111187. eCollection 2014. PLoS One. 2014. PMID: 25347823 Free PMC article. - Spatial and functional organization of mitochondrial protein network.
Yang JS, Kim J, Park S, Jeon J, Shin YE, Kim S. Yang JS, et al. Sci Rep. 2013;3:1403. doi: 10.1038/srep01403. Sci Rep. 2013. PMID: 23466738 Free PMC article. - MitoMiner: a data warehouse for mitochondrial proteomics data.
Smith AC, Blackshaw JA, Robinson AJ. Smith AC, et al. Nucleic Acids Res. 2012 Jan;40(Database issue):D1160-7. doi: 10.1093/nar/gkr1101. Epub 2011 Nov 24. Nucleic Acids Res. 2012. PMID: 22121219 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous