BAC to the future! or oligonucleotides: a perspective for micro array comparative genomic hybridization (array CGH) - PubMed (original) (raw)
Review
. 2006 Jan 26;34(2):445-50.
doi: 10.1093/nar/gkj456. Print 2006.
Affiliations
- PMID: 16439806
- PMCID: PMC1356528
- DOI: 10.1093/nar/gkj456
Review
BAC to the future! or oligonucleotides: a perspective for micro array comparative genomic hybridization (array CGH)
Bauke Ylstra et al. Nucleic Acids Res. 2006.
Abstract
The array CGH technique (Array Comparative Genome Hybridization) has been developed to detect chromosomal copy number changes on a genome-wide and/or high-resolution scale. It is used in human genetics and oncology, with great promise for clinical application. Until recently primarily PCR amplified bacterial artificial chromosomes (BACs) or cDNAs have been spotted as elements on the array. The large-scale DNA isolations or PCR amplifications of the large-insert clones necessary for manufacturing the arrays are elaborate and time-consuming. Lack of a high-resolution highly sensitive (commercial) alternative has undoubtedly hindered the implementation of array CGH in research and diagnostics. Recently, synthetic oligonucleotides as arrayed elements have been introduced as an alternative substrate for array CGH, both by academic institutions as well as by commercial providers. Oligonucleotide libraries or ready-made arrays can be bought off-the-shelf saving considerable time and efforts. For RNA expression profiling, we have seen a gradual transition from in-house printed cDNA-based expression arrays to oligonucleotide arrays and we expect a similar transition for array CGH. This review compares the different platforms and will attempt to shine a light on the 'BAC to the future' of the array CGH technique.
Figures
Figure 1
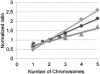
Relation between theoretical and measured chromosomal copy number changes for several different platforms. Number of chromosomes on the horizontal axis and the dye normalized experimental ratios on the vertical axis. Agilent custom oaCGH (25), speckled gray line with diamonds, slope 0.53; BAC arrays (3), thin line with dots, slope 0.37; in-house printed oaCGH (14), short dashes line with plusses, slope 0.28; Agilent oaCGH on expression arrays (25), dark gray line with triangles slope 0.21. A single chromosomal copy gain in a diploid background theoretically shifts the ratio from 1 to 1.5 (two chromosomes/two chromosomes versus three chromosomes/two chromosomes). To detect a single copy gain in a diploid background, the total variance should therefore be smaller than 0.25 so that there is no overlap between normal and gain. The copy number values and slope for the Agilent CGH platform (25) is nearly identical to the theoretical values and slope. However, the variance for one or even three array elements combined is too high to unequivocally call a gain (Table 1). The relation between the theoretical and measured chromosomal copy number change for a given BAC array is slightly compressed, but the variance is maximally 0.18 (Table 1). This makes it possible to make a call on a single arrayed BAC element. For the in-house printed oaCGH platform the relation between theoretical and measured chromosomal copy number is also not ideal; however, three oligonucleotides are still sufficient to call a single copy number change, since the elements have a limited variance (Table 1). The ROMA or NimbleGen platforms could not be included, since raw log2 ratios were not available. The Affymetrix (22) platform was not included since it is a single channel array and this figure presents normalized ratios.
Similar articles
- Human and mouse oligonucleotide-based array CGH.
van den Ijssel P, Tijssen M, Chin SF, Eijk P, Carvalho B, Hopmans E, Holstege H, Bangarusamy DK, Jonkers J, Meijer GA, Caldas C, Ylstra B. van den Ijssel P, et al. Nucleic Acids Res. 2005 Dec 16;33(22):e192. doi: 10.1093/nar/gni191. Nucleic Acids Res. 2005. PMID: 16361265 Free PMC article. - BAC-based PCR fragment microarray: high-resolution detection of chromosomal deletion and duplication breakpoints.
Ren H, Francis W, Boys A, Chueh AC, Wong N, La P, Wong LH, Ryan J, Slater HR, Choo KH. Ren H, et al. Hum Mutat. 2005 May;25(5):476-82. doi: 10.1002/humu.20164. Hum Mutat. 2005. PMID: 15832308 - A cytogeneticist's perspective on genomic microarrays.
Shaffer LG, Bejjani BA. Shaffer LG, et al. Hum Reprod Update. 2004 May-Jun;10(3):221-6. doi: 10.1093/humupd/dmh022. Hum Reprod Update. 2004. PMID: 15140869 Review. - Combined array-comparative genomic hybridization and single-nucleotide polymorphism-loss of heterozygosity analysis reveals complex genetic alterations in cervical cancer.
Kloth JN, Oosting J, van Wezel T, Szuhai K, Knijnenburg J, Gorter A, Kenter GG, Fleuren GJ, Jordanova ES. Kloth JN, et al. BMC Genomics. 2007 Feb 20;8:53. doi: 10.1186/1471-2164-8-53. BMC Genomics. 2007. PMID: 17311676 Free PMC article. - Microarray-based comparative genomic hybridization and its applications in human genetics.
Oostlander AE, Meijer GA, Ylstra B. Oostlander AE, et al. Clin Genet. 2004 Dec;66(6):488-95. doi: 10.1111/j.1399-0004.2004.00322.x. Clin Genet. 2004. PMID: 15521975 Review.
Cited by
- Copy number variants in a population-based investigation of Klippel-Trenaunay syndrome.
Dimopoulos A, Sicko RJ, Kay DM, Rigler SL, Fan R, Romitti PA, Browne ML, Druschel CM, Caggana M, Brody LC, Mills JL. Dimopoulos A, et al. Am J Med Genet A. 2017 Feb;173(2):352-359. doi: 10.1002/ajmg.a.37868. Epub 2016 Nov 30. Am J Med Genet A. 2017. PMID: 27901321 Free PMC article. - A Robust Protocol for Using Multiplexed Droplet Digital PCR to Quantify Somatic Copy Number Alterations in Clinical Tissue Specimens.
Hughesman CB, Lu XJ, Liu KY, Zhu Y, Poh CF, Haynes C. Hughesman CB, et al. PLoS One. 2016 Aug 18;11(8):e0161274. doi: 10.1371/journal.pone.0161274. eCollection 2016. PLoS One. 2016. PMID: 27537682 Free PMC article. - A nCounter CNV Assay to Detect HER2 Amplification: A Correlation Study with Immunohistochemistry and In Situ Hybridization in Advanced Gastric Cancer.
Ahn S, Hong M, Van Vrancken M, Lyou YJ, Kim ST, Park SH, Kang WK, Park YS, Jung SH, Woo M, Lee J, Kim KM. Ahn S, et al. Mol Diagn Ther. 2016 Aug;20(4):375-83. doi: 10.1007/s40291-016-0205-4. Mol Diagn Ther. 2016. PMID: 27179810 - Penalized weighted low-rank approximation for robust recovery of recurrent copy number variations.
Gao X. Gao X. BMC Bioinformatics. 2015 Dec 10;16:407. doi: 10.1186/s12859-015-0835-2. BMC Bioinformatics. 2015. PMID: 26652207 Free PMC article. - aCGH-MAS: analysis of aCGH by means of multiagent system.
De Paz JF, Benito R, Bajo J, Rodríguez AE, Abáigar M. De Paz JF, et al. Biomed Res Int. 2015;2015:194624. doi: 10.1155/2015/194624. Epub 2015 Mar 22. Biomed Res Int. 2015. PMID: 25874203 Free PMC article. Review.
References
- Kallioniemi A., Kallioniemi O.P., Sudar D., Rutovitz D., Gray J.W., Waldman F., Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. - PubMed
- Pollack J.R., Perou C.M., Alizadeh A.A., Eisen M.B., Pergamenschikov A., Williams C.F., Jeffrey S.S., Botstein D., Brown P.O. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nature Genet. 1999;23:41–46. - PubMed
- Pinkel D., Segraves R., Sudar D., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nature Genet. 1998;20:207–211. - PubMed
- Solinas-Toldo S., Lampel S., Stilgenbauer S., Nickolenko J., Benner A., Dohner H., Cremer T., Lichter P. Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer. 1997;20:399–407. - PubMed
- Pinkel D., Albertson D.G. Array comparative genomic hybridization and its applications in cancer. Nature Genet. 2005;37(Suppl.):S11–S17. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources