Transposable elements have contributed to thousands of human proteins - PubMed (original) (raw)
Transposable elements have contributed to thousands of human proteins
Roy Britten. Proc Natl Acad Sci U S A. 2006.
Abstract
This is a report of many distant but significant protein sequence relationships between human proteins and transposable elements (TEs). The libraries of human repeated sequences contain the DNA sequences of many TEs. These were translated in all reading frames, ignoring stop codons, and were used as amino acid sequence probes to search with BLASTP for similar sequences in a library of 25,193 human proteins. The probes show regions of significant amino acid sequence similarity to 1,950 different human genes, with an expectation of <10(-3). In comparison with previous REPEATMASKER (Institute for Systems Biology, Seattle) studies, these probes detect many more TE sequences in more human coding sequences with greater length than previous work using DNA sequences. If the criterion is opened, very many matches are found occurring on 4,653 different genes after correction for the number seen with random amino acid sequence probes. The processes that led to these extensive sets of sequence relationships between TEs and coding sequences of human genes have been a major source of variation and novel genes during evolution. This paper lists the number of sequence similarities seen by amino acid sequence comparison, which is surely an underestimate of the actual number of significant relationships. It appears that many of these are the result of past events of duplication of genes or gene regions, rather than a direct result of TE insertion. This report of observable relationships leaves to the future the functional implications as well as the detection of the events of TE insertion.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
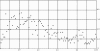
Fig. 1.
The number of the 1,950 matches vs. coverage. Horizontally is the fraction of the length of the gene included in all of the matches to the amino acid sequence probes in 1% intervals. Vertically is the number of genes in each interval. Many different TEs contribute to the coverage of individual proteins.
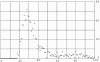
Fig. 2.
The number of matches vs. maximum percent match. Vertically is the number of matches in each interval of 1%.
Similar articles
- Almost all human genes resulted from ancient duplication.
Britten RJ. Britten RJ. Proc Natl Acad Sci U S A. 2006 Dec 12;103(50):19027-32. doi: 10.1073/pnas.0608796103. Epub 2006 Dec 4. Proc Natl Acad Sci U S A. 2006. PMID: 17146051 Free PMC article. - The majority of human genes have regions repeated in other human genes.
Britten RJ. Britten RJ. Proc Natl Acad Sci U S A. 2005 Apr 12;102(15):5466-70. doi: 10.1073/pnas.0501008102. Epub 2005 Mar 31. Proc Natl Acad Sci U S A. 2005. PMID: 15802472 Free PMC article. - Fungal transposable elements and genome evolution.
Daboussi MJ. Daboussi MJ. Genetica. 1997;100(1-3):253-60. Genetica. 1997. PMID: 9440278 Review. - Exaptation of protein coding sequences from transposable elements.
Bowen NJ, Jordan IK. Bowen NJ, et al. Genome Dyn. 2007;3:147-162. doi: 10.1159/000107609. Genome Dyn. 2007. PMID: 18753790 Review.
Cited by
- Evolutionary rate of human tissue-specific genes are related with transposable element insertions.
Jin P, Qin S, Chen X, Song Y, Li-Ling J, Xu X, Ma F. Jin P, et al. Genetica. 2012 Dec;140(10-12):513-23. doi: 10.1007/s10709-013-9700-2. Epub 2013 Jan 22. Genetica. 2012. PMID: 23337972 - HERV-E-mediated modulation of PLA2G4A transcription in urothelial carcinoma.
Gosenca D, Gabriel U, Steidler A, Mayer J, Diem O, Erben P, Fabarius A, Leib-Mösch C, Hofmann WK, Seifarth W. Gosenca D, et al. PLoS One. 2012;7(11):e49341. doi: 10.1371/journal.pone.0049341. Epub 2012 Nov 7. PLoS One. 2012. PMID: 23145155 Free PMC article. - Biochemical Properties and Physiological Functions of pLG72: Twenty Years of Investigations.
Murtas G, Pollegioni L, Molla G, Sacchi S. Murtas G, et al. Biomolecules. 2022 Jun 20;12(6):858. doi: 10.3390/biom12060858. Biomolecules. 2022. PMID: 35740983 Free PMC article. Review. - Structure prediction and analysis of DNA transposon and LINE retrotransposon proteins.
Abrusán G, Zhang Y, Szilágyi A. Abrusán G, et al. J Biol Chem. 2013 May 31;288(22):16127-38. doi: 10.1074/jbc.M113.451500. Epub 2013 Mar 25. J Biol Chem. 2013. PMID: 23530042 Free PMC article. - Mobile DNA and evolution in the 21st century.
Shapiro JA. Shapiro JA. Mob DNA. 2010 Jan 25;1(1):4. doi: 10.1186/1759-8753-1-4. Mob DNA. 2010. PMID: 20226073 Free PMC article.
References
- Brownell E., Mittereder N., Rice N. R. Oncogene. 1989;4:935–942. - PubMed
- Smit A. F. Genet. Dev. 1999;9:657–663. - PubMed
- Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Nature. 2001;409:860–921. - PubMed
- Nekrutenko A., Li W. H. Trends Genet. 2001;17:619–621. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous