CAND1-mediated substrate adaptor recycling is required for efficient repression of Nrf2 by Keap1 - PubMed (original) (raw)
CAND1-mediated substrate adaptor recycling is required for efficient repression of Nrf2 by Keap1
Shih-Ching Lo et al. Mol Cell Biol. 2006 Feb.
Abstract
The bZIP transcription factor Nrf2 controls a genetic program that protects cells from oxidative damage and maintains cellular redox homeostasis. Keap1, a BTB-Kelch protein, is the major upstream regulator of Nrf2. Keap1 functions as a substrate adaptor protein for a Cul3-dependent E3 ubiquitin ligase complex to repress steady-state levels of Nrf2 and Nrf2-dependent transcription. Cullin-dependent ubiquitin ligase complexes have been proposed to undergo dynamic cycles of assembly and disassembly that enable substrate adaptor exchange or recycling. In this report, we have characterized the importance of substrate adaptor recycling for regulation of Keap1-mediated repression of Nrf2. Association of Keap1 with Cul3 was decreased by ectopic expression of CAND1 and was increased by small interfering RNA (siRNA)-mediated knockdown of CAND1. However, both ectopic overexpression and siRNA-mediated knockdown of CAND1 decreased the ability of Keap1 to target Nrf2 for ubiquitin-dependent degradation, resulting in stabilization of Nrf2 and activation of Nrf2-dependent gene expression. Neddylation of Cul3 on Lys 712 is required for Keap1-dependent ubiquitination of Nrf2 in vivo. However, the K712R mutant Cul3 molecule, which is not neddylated, can still assemble with Keap1 into a functional ubiquitin ligase complex in vitro. These results provide support for a model in which substrate adaptor recycling is required for efficient substrate ubiquitination by cullin-dependent E3 ubiquitin ligase complexes.
Figures
FIG. 1.
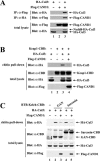
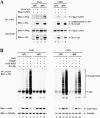
(A) Thirty-five-millimeter-diameter dishes of COS1 cells were transfected with 0.5 μg each of expression vectors for HA-Cul3 and Flag-CAND1 as indicated. Total cell lysates were analyzed by immunoblotting with anti-Flag (α-Flag) and α-HA antibodies (bottom two panels). α-Flag immunoprecipitates (IP) were subjected to immunoblot analysis using α-HA antibodies (top panel). α-HA IP were subjected to immunoblot analysis using α-Flag antibodies (second panel from the top). (B) Thirty-five-millimeter-diameter dishes of COS1 cells were transfected with 0.33 μg each of expression vectors for Keap1-CBD and HA-Cul3 as indicated. The Flag-CAND1 expression vector was either omitted (lanes 1 to 3) or included (lane 4). Total cell lysates were analyzed by immunoblotting with α-CBD, α-Flag, and α-HA antibodies (bottom three panels). The lysates were incubated with chitin beads, pelleted by centrifugation (3,000 × g), and washed three times in lysis buffer. Proteins that remained associated with the chitin beads were analyzed by immunoblotting with α-HA antibodies (top panel). (C) Thirty-five-millimeter-diameter dishes of COS1 cells were transfected with expression vectors for HA-Cul3 (lanes 1 to 5) and GAN-CBD (lanes 2 and 3) or sarcosin-CBD (lanes 4 and 5). The Flag-CAND1 expression vector was either omitted (lanes 1, 2, and 4) or included (lanes 3 and 5). Cell lysates were analyzed by immunoblotting with the indicated antibodies (bottom three panels) or incubated with chitin beads. Proteins that remained associated with the chitin beads after extensive washing were analyzed by immunoblotting with α-HA antibodies (top panel).
FIG. 2.
(A) Thirty-five-millimeter-diameter dishes of MDA-MB-231 cells were transfected with expression vectors for HA-Ub (0.4 μg), Gal4-Neh2 (0.4 μg, lanes 2 to 7), Keap1 (0.1 μg, lanes 3 to 7), Cul3 (0.1 μg, lanes 3 to 7), and CAND1 (from 0.0125 to 0.1 μg, lanes 4 to 7). The transfected cells were treated with MG132 for 5 h prior to cell lysis. Total cell lysates were analyzed by immunoblotting with anti-Gal4 (α-Gal4) antibodies (bottom panel). α-Gal4 immunoprecipitates (IP) were analyzed by immunoblotting with α-HA antibodies (top panel). IgG, immunoglobulin G. (B) Twenty-four-well plates of MDA-MB-231 cells were transfected with expression vectors for HA-Nrf2 (0.18 μg), Keap1 (0.018 μg, lanes 2 to 5), Cul3 (0.07 μg, lanes 2 to 5), and CAND1 (from 0.035 to 0.14 μg, lanes 3 to 5). Total cell lysates were subjected to immunoblot analysis with α-HA (top panel) and α-tubulin (bottom panel) antibodies. (C) Twenty-four-well plates of HeLa cells were transfected with expression vectors for HA-Nrf2 (0.18 μg), Keap1 (0.018 μg, lanes 2 to 4), and Flag-CAND1 (0.035 to 0.14 μg, lanes 3 and 4). Total cell lysates were subjected to immunoblot analysis with α-HA, α-Keap1, α-Flag, and α-tubulin antibodies as indicated.
FIG. 3.
(A) Thirty-five-millimeter-diameter dishes of HeLa cells were transfected with control (lane 4) or anti-CAND1 (α-CAND1) (lane 3) siRNA nucleotides (300 nM), allowed to recover for 24 h, and transfected with 0.5 μg each of expression vectors for HA-Cul3 and Keap1-CBD (lanes 2 to 4). Cell lysates were collected after an additional 24 h and immunoblotted with the indicated antibodies (bottom four panels) or incubated with chitin beads. Proteins that remained bound to the chitin beads after extensive washing were analyzed with the indicated antibodies (top two panels). (B) HeLa cells were transfected with control (lane 4) or α-CAND1 (lane 3) siRNA nucleotides (100 nM), allowed to recover for 24 h, and transfected with expression vectors for HA-Ub, Keap1, and Cul3 as indicated. Cell lysates were collected after an additional 24 h and immunoblotted with the indicated antibodies (bottom three panels). α-Keap1 immunoprecipitates (IP) were analyzed by immunoblotting with α-HA antibodies (top panel). IgG, immunoglobulin G. (C) Twenty-four-well plates of HeLa cells were transfected with α-Nrf2 (lane 3), α-Keap1 (lane 4), or α-CAND1 (lane 5) siRNA nucleotides (50 nM), allowed to recover for 24 h, and then transfected with 0.2 μg each of expression vectors for HA-Nrf2 and Keap1 (lanes 2 to 5). Total cell lysates were collected and subjected to immunoblot analysis with the indicated antibodies. (D) Thirty-five-millimeter-diameter dishes of HeLa cells were first mock transfected (lanes 1 and 3) or transfected with α-CAND1 siRNA nucleotides (300 nM), allowed to recover for 24 h, and then transfected with 0.5 μg each of expression vectors for HA-Nrf2 and Keap1-CBD. The transfected cells were either untreated (lanes 1 and 2) or treated with MG132 for 5 h prior to cell lysis. The lysates were incubated with chitin beads, and levels of HA-Nrf2 proteins that remained bound to the chitin beads were determined by immunoblotting with α-HA antibodies (top panel). Total lysates were analyzed by immunoblotting with the indicated antibodies (bottom three panels).
FIG. 4.
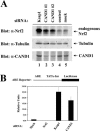
(A) Twenty-four-well plates of HeLa cells were transfected with 100 nM of control siRNA (lane 4) or siRNA nucleotides targeting Keap1 (lane 1) or CAND1 at different regions in CAND1 mRNA (lanes 2 and 3). Levels of endogenous Nrf2 were determined by immunoblot analysis with anti-Nrf2 (α-Nrf2) antibodies (top panel). Levels of CAND1 and tubulin were analyzed by immunoblotting with the indicated antibodies (bottom two panels). (B) Twenty-four-well plates of HeLa cells were transfected with α-Nrf2, α-Keap1, or α-CAND1 siRNA nucleotides (300 nM) as indicated and with an ARE-dependent firefly luciferase reporter gene construct (100 ng). A plasmid encoding Renilla luciferase (10 ng) was included as a control for transfection efficiency. The data shown represent the means and standard deviations of results from three independent experiments.
FIG. 5.
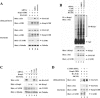
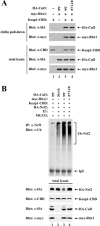
(A) Sixty-millimeter-diameter dishes of ts41 cells or wild-type CHO cells were cotransfected with 1.0 μg each of expression vectors for HA-Cul3 and Flag-CAND1 as indicated. The transfected cells were either kept at 34°C (lanes 1, 2, 5, and 6) or shifted to 40°C (lanes 3, 4, 7, and 8) for 24 h. Total cell lysates were analyzed by immunoblotting with anti-Flag (α-Flag) and α-HA antibodies (bottom two panels). α-HA immunoprecipitates (IP) were subjected to immunoblot analysis using α-Flag and α-HA antibodies (top two panels). (B) Sixty-millimeter-diameter dishes of ts41 cells or wild-type CHO cells were cotransfected with expression vectors for HA-Ub (0.7 μg), Gal4-Neh2 (0.8 μg), Keap1 (0.2 μg), and Cul3 (0.3 μg) as indicated. The transfected cells were either kept at 34°C (lanes 1 to 4 and 9 to 12) or shifted to 40°C (lanes 5 to 8 and 13 to 16) for 24 h, and all were treated with MG132 for 2.5 h prior to cell lysis. Total cell lysates were analyzed by immunoblotting with α-Gal4 and α-tubulin antibodies (bottom two panels). α-Gal4 IP were analyzed by immunoblotting with α-HA antibodies (top panel). IgG, immunoglobulin G.
FIG. 6.
(A) Sequence alignment of neddylation sites among five members of the cullin superfamily is shown. The conserved Lys residue for Nedd8 conjugation in the cullin proteins is indicated above the alignment by an arrow (39). Conserved residues in this region are indicated below the alignment by asterisks. Residues in this region of Cul1 that make direct contacts with CAND1 are indicated above the alignment by carets (17). The conserved residues that were substituted with alanine or arginine in the Cul3 mutant proteins utilized in this report are illustrated. Hs., Homo sapiens. (B) Thirty-five-millimeter-diameter dishes of COS1 cells were cotransfected with 0.5 μg each of expression vectors for Flag-CAND1 and the mutant or wild-type (WT) HA-Cul3 proteins as indicated. Cell lysates were collected in the presence of 10 mM NEM and analyzed by immunoblotting with anti-Flag (α-Flag) and α-HA antibodies (bottom two panels). α-HA immunoprecipitates (IP) were subjected to immunoblot analysis using α-Flag antibodies (top panel). (C) Thirty-five-millimeter-diameter dishes of MDA-MB-231 cells were transfected with expression vectors for HA-Ub (0.4 μg), Gal4-Neh2 (0.4 μg, lanes 2 to 11), Keap1 (0.1 μg, lanes 3 to 11), and the WT or mutant Cul3 proteins (0.1 μg, lanes 4 to 11). The transfected cells were treated with MG132 for 2.5 h prior to cell lysis. α-Gal4 IP from cell lysates were analyzed by immunoblotting with α-HA antibodies. IgG, immunoglobulin G. (D) Twenty-four-well plates of MDA-MB-231 cells were transfected with expression vectors for HA-Nrf2 (0.18 μg), myc-Rbx1 (0.11 μg, lanes 2 to 10), Keap1 (0.018 μg, lanes 2 to 10), and the WT or mutant Cul3 proteins (0.11 μg, lanes 3 to 10). Total cell lysates were subjected to immunoblot analysis with α-HA (top and middle panels) and α-tubulin (bottom panel) antibodies.
FIG. 7.
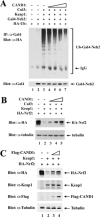
(A) Thirty-five-millimeter-diameter dishes of COS1 cells were cotransfected with expression vectors for Keap1-CBD (0.3 μg, lanes 1, 3, and 4), myc-Rbx1 (0.1 μg, lanes 2 to 4), and the wild-type (WT) or mutant HA-Cul3 proteins (0.3 μg, lanes 2 to 4) as indicated. Cell lysates were analyzed by immunoblotting with the indicated antibodies (bottom three panels) or incubated with chitin beads. Proteins that remained bound to the chitin beads after extensive washing were analyzed by immunoblotting with the indicated antibodies (top two panels). (B) Sixty-millimeter-diameter dishes of COS1 cells were transfected with 0.5 μg each of the expression vectors for HA-Nrf2, Keap1-CBD, myc-Rbx1, and the WT or mutant HA-Cul3 proteins, as indicated in lanes 1 to 4. The transfected cells were treated with MG132 for 5 h prior to cell lysis. Lysates from three 60-mm-diameter dishes were pooled for each sample. One percent of the cell lysates were analyzed by immunoblotting with the indicated antibodies (bottom four panels), and the rest of the lysates were incubated with chitin beads. After washing, the chitin beads were incubated with E1, E2-UbcH5a, ubiquitin, and ATP. The E1 enzyme was omitted from one sample (lane 1). Subsequently, the chitin beads were pelleted and washed, and proteins that were eluted from the beads after boiling under denaturing conditions were immunoprecipitated with anti-Nrf2 (α-Nrf2) antibodies and then analyzed by immunoblotting with antiubiquitin antibodies (top panel). IP, immunoprecipitate; IgG, immunoglobulin G.
Similar articles
- Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin-ring ubiquitin ligases.
Villeneuve NF, Lau A, Zhang DD. Villeneuve NF, et al. Antioxid Redox Signal. 2010 Dec 1;13(11):1699-712. doi: 10.1089/ars.2010.3211. Epub 2010 Aug 14. Antioxid Redox Signal. 2010. PMID: 20486766 Free PMC article. Review. - Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex.
Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Zhang DD, et al. Mol Cell Biol. 2004 Dec;24(24):10941-53. doi: 10.1128/MCB.24.24.10941-10953.2004. Mol Cell Biol. 2004. PMID: 15572695 Free PMC article. - Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2.
Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Kobayashi A, et al. Mol Cell Biol. 2004 Aug;24(16):7130-9. doi: 10.1128/MCB.24.16.7130-7139.2004. Mol Cell Biol. 2004. PMID: 15282312 Free PMC article. - BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase.
Furukawa M, Xiong Y. Furukawa M, et al. Mol Cell Biol. 2005 Jan;25(1):162-71. doi: 10.1128/MCB.25.1.162-171.2005. Mol Cell Biol. 2005. PMID: 15601839 Free PMC article. - CRL3s: The BTB-CUL3-RING E3 Ubiquitin Ligases.
Wang P, Song J, Ye D. Wang P, et al. Adv Exp Med Biol. 2020;1217:211-223. doi: 10.1007/978-981-15-1025-0_13. Adv Exp Med Biol. 2020. PMID: 31898230 Review.
Cited by
- Activation of Nrf2 by arsenite and monomethylarsonous acid is independent of Keap1-C151: enhanced Keap1-Cul3 interaction.
Wang XJ, Sun Z, Chen W, Li Y, Villeneuve NF, Zhang DD. Wang XJ, et al. Toxicol Appl Pharmacol. 2008 Aug 1;230(3):383-9. doi: 10.1016/j.taap.2008.03.003. Epub 2008 Mar 12. Toxicol Appl Pharmacol. 2008. PMID: 18417180 Free PMC article. - The COP9 signalosome, cullin 3 and Keap1 supercomplex regulates CHOP stability and adipogenesis.
Huang X, Ordemann J, Müller JM, Dubiel W. Huang X, et al. Biol Open. 2012 Aug 15;1(8):705-10. doi: 10.1242/bio.20121875. Epub 2012 Jun 12. Biol Open. 2012. PMID: 23213463 Free PMC article. - Targeting neddylation and sumoylation in chemoresistant triple negative breast cancer.
Powell RT, Rinkenbaugh AL, Guo L, Cai S, Shao J, Zhou X, Zhang X, Jeter-Jones S, Fu C, Qi Y, Baameur Hancock F, White JB, Stephan C, Davies PJ, Moulder S, Symmans WF, Chang JT, Piwnica-Worms H. Powell RT, et al. NPJ Breast Cancer. 2024 May 27;10(1):37. doi: 10.1038/s41523-024-00644-4. NPJ Breast Cancer. 2024. PMID: 38802426 Free PMC article. - Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin-ring ubiquitin ligases.
Villeneuve NF, Lau A, Zhang DD. Villeneuve NF, et al. Antioxid Redox Signal. 2010 Dec 1;13(11):1699-712. doi: 10.1089/ars.2010.3211. Epub 2010 Aug 14. Antioxid Redox Signal. 2010. PMID: 20486766 Free PMC article. Review. - Neuroprotective Effects and Therapeutic Potential of the Citrus Flavonoid Hesperetin in Neurodegenerative Diseases.
Evans JA, Mendonca P, Soliman KFA. Evans JA, et al. Nutrients. 2022 May 26;14(11):2228. doi: 10.3390/nu14112228. Nutrients. 2022. PMID: 35684025 Free PMC article. Review.
References
- Ames, B. N., and M. K. Shigenaga. 1993. DNA and free radicals, p. 1-15. In B. Halliwell and O. I. Aruoma (ed.), Oxidants are a major contributor to cancer and aging. Ellis Horwood, New York, N.Y.
- Ceconi, C., A. Boraso, A. Cargnoni, and R. Ferrari. 2003. Oxidative stress in cardiovascular disease: myth or fact? Arch. Biochem. Biophys. 420:217-221. - PubMed
- Chen, Y., D. L. McPhie, J. Hirschberg, and R. L. Neve. 2000. The amyloid precursor protein-binding protein APP-BP1 drives the cell cycle through the S-M checkpoint and causes apoptosis in neurons. J. Biol. Chem. 275:8929-8935. - PubMed
- Cope, G. A., and R. J. Deshaies. 2003. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 114:663-671. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases