Building of the tetraspanin web: distinct structural domains of CD81 function in different cellular compartments - PubMed (original) (raw)
Building of the tetraspanin web: distinct structural domains of CD81 function in different cellular compartments
Tsipi Shoham et al. Mol Cell Biol. 2006 Feb.
Abstract
The tetraspanin web is composed of a network of tetraspanins and their partner proteins that facilitate cellular interactions and fusion events by an unknown mechanism. Our aim was to unravel the web partnership between the tetraspanin CD81 and CD19, a cell surface signaling molecule in B lymphocytes. We found that CD81 plays multiple roles in the processing, intracellular trafficking, and membrane functions of CD19. Surprisingly, these different roles are embodied in distinct CD81 domains, which function in the different cellular compartments: the N-terminal tail of CD81 has an effect on the glycosylation of CD19; the first transmembrane domain of CD81 is sufficient to support the exit of CD19 from the endoplasmic reticulum, although the large extracellular loop (LEL) of CD81 associates physically with CD19 early during biosynthesis; and finally, the TM2 and TM3 domains of CD81 play a role in the transmission of signals initiated upon engagement of the LEL. The participation of distinct CD81 domains in varied functions may explain the pleiotropic effects of CD81 within the tetraspanin web.
Figures
FIG. 1.
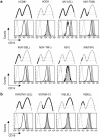
The first transmembrane domain (TM1) of hCD81 is sufficient for normal surface expression of CD19. A panel of retroviruses encoding hCD81 (boldface lines), hCD9 (dotted lines), or chimeric hCD81/hCD9 molecules were used to infect the _Cd81_−/− early B-cell line 1C8. Roman numerals represent laboratory designations for the chimeric molecules, and the CD81 region in each chimera is in parentheses. Cells were infected with the indicated tetraspanin-IRES-GFP retroviruses and stained with an anti-CD19 MAb (filled histograms). In parallel, CD19 expression was analyzed in cells infected at the same time with a control GFP retrovirus (boldface-line histograms). Each of the panels compares CD19 expression in the transduced cells (gated GFP positive). Cells were also stained with an isotype control MAbs (dotted-line histograms). The infected cells express equivalent levels of the transduced molecules (see Fig. S3 in the supplemental material). Only chimeras that contain the CD81 TM1 domain express normal levels of CD19, which are two to threefold higher than the levels expressed in 1C8 cells. (a) Replacement of CD81 domains from the C terminus with the corresponding CD9 sequence; (b) replacement of CD81 domains from the N terminus with the corresponding CD9 sequence. The data represent two or three independent flow cytometry analyses for each chimera.
FIG. 2.
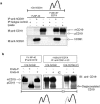
CD81 affects intracellular processing of CD19. Analysis of CD19 glycoforms expressed in the _Cd81_−/− cell line (1C8) and in subclones expressing hCD81 or chimeric tetraspanin molecules. The mature (mCD19) (solid arrows) and the precursor (pCD19) (dashed arrow) CD19 glycoforms were detected by immunoblotting with rabbit anti-CD19 Ab. (a) Cells lacking CD81 (1C8) accumulate smaller amounts of slow-migrating mCD19 and larger amounts of pCD19 than CD81-expressing cells (1C8/hCD81); the data represent one of three analyses. (b) Sensitivity of the CD19 glycoforms to digestion by endo-F and endo-H. Immunoprecipitated (IP) CD19 molecules were digested with endo-H or endo-F, as indicated, and detected by immunoblotting (IB). The CD19 molecules in both cell lines are equally sensitive to endo-F digestion, and the deglycosylated CD19 molecules migrate similarly. The difference in the migration of undigested mCD19 molecules in 1C8 and 1C8/hCD81 cells is therefore due to a variation in glycosylation. These data represent one of two analyses. (c) Analysis of subclones expressing hCD81 and chimeric molecules. The mCD19 molecules are more abundant in cells expressing the TM1 domains of CD81 [hCD81, II(N′-TM3), IV(N′-TM1), and XIII(TM1)], whereas pCD19 molecules are more abundant in cells that do not contain TM1 [VII(LEL) and VI(TM2-C′)]. The membrane was reblotted with an antiactin Ab (lower panel). (d) The ratios between pCD19 and mCD19 in each lane were determined by densitometry. Subclones that support optimal expression of CD19 (Fig. 1) have low pCD19/mCD19 ratios (white bars), whereas subclones that do not support CD19 expression have higher pCD19/mCD19 ratio (gray bars). The data represent a summary of four independent experiments; error bars indicate SD.
FIG. 3.
CD81 facilitates the exit of CD19 from the ER. (a) Pulse-chase analysis of CD19 in cells labeled for 30 min with [35S]methionine and [35S]cysteine and then chased for the indicated time periods. Labeled cells were lysed, followed by immunoprecipitation (IP) with an anti-CD19 MAb (1D3) or isotype control MAb (IC). In the presence of CD81 (1C8/hCD81 cells, left panel), mCD19 molecules (solid arrow) can already be seen at the end of the pulse period (0-min chase), and most of the pCD19 molecules (dashed arrow) are chased within 1 h. In the absence of CD81 (1C8 cells, right panel) mCD19 molecules can be detected only after 1 h of chase, while pCD19 molecules can still be seen even after 2 h of chase. The open arrowheads point to nonspecific bands, immunoprecipitated by the isotype control MAb. (b) The amounts of mCD19 and pCD19 in each lane were determined by densitometry and are presented as ratios of these glycoforms. These data represent three independent experiments.
FIG. 4.
CD81 associates with both the precursor and the mature CD19 glycoforms. 1C8/hCD81 cells were lysed in the indicated lysis buffer, immunoprecipitated (IP), and immunoblotted (IB) with the indicated MAbs. Lanes contain either 10 μl of cell lysates or immunoprecipitated molecules from 400 μl of lysate. (a) The anti-CD81 MAb coprecipitates both pCD19 and mCD19 under mild (Brij 97), but not harsh (NP-40) lysis conditions. The data represent two independent experiments. (b) Cell lysates of 1C8/hCD81 or 1C8/VI(TM2-C′) cells, immunoprecipitated with the indicated MAbs, were digested by either endo-H or endo-F, and CD19 molecules were immunoblotted. The anti-hCD81 MAb coprecipitates pCD19 molecules, which are sensitive to endo-H digestion and migrate similarly to the endo-F-deglycosylated CD19. IC, isotype control.
FIG. 5.
CD81 LEL augments the association with CD19. The indicated subclones were lysed as indicated, and 400 μl of each lysate was immunoprecipitated (IP) with anti-CD19 (1D3) or directly loaded (10 μl per lane). The coprecipitated tetraspanins were immunoblotted (IB) with (a) anti-hCD81 [chimera VII(LEL)] or (b) anti-hCD9 [(chimera XIII(TM1) and 1C8/hCD9)] (upper panels). The anti-CD19 Ab coprecipitates a larger fraction of chimera VII(LEL) than chimera XIII(TM1) or the whole hCD9 molecules. The ratio of CD19-precipitated tetraspanin molecules to the total amount of CD19 is the highest in chimera VII(LEL), as shown by reblotting of the membranes with anti-CD19 Abs (lower panels). The data represent two independent experiments.
FIG. 6.
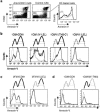
Engagement of CD81 on the cell surface increases annexin V binding and requires the CD81 LEL and TM2-TM3 domains. (a) 1C8/hCD81 cells were incubated with the biotinylated anti-hCD81 MAb 5A6 (left panel) or with a biotinylated isotype control MAb (middle panel) and then cross-linked by streptavidin. Cross-linking of CD81 (solid line) induced an increase in annexin V binding in live cells (7-amino-actinomycin D [7AAD] negative, R3-gated cells) in comparison to treatment with the control MAb (dotted line), as shown in the histograms (right panel). (b) 1C8 cells expressing the indicated chimera were stimulated as for panel a. Live cells (GFP positive) (see Fig. S5a in the supplemental material) were gated and analyzed for annexin V binding. All analyzed cells express the hCD81 LEL and bind the 5A6 MAb (see Fig. S3 in the supplemental material); however, only cells that contain the CD81 TM2 and TM3 domains show an increase in annexin V binding in response to cross-linking by the antibody. Similar results were obtained by cross-linking using unconjugated MAbs and a secondary anti-mouse Ig Ab (data not shown). These data represent six independent experiments using cells purified from three independent retroviral infections. (c) The Cd81 +/+ early B-cell line 2F3 was infected by retroviruses encoding hCD81 or chimera VII(LEL) (see Fig. S5b in the supplemental material). Infected cells (GFP positive) were sorted and analyzed for annexin V binding following cross-linking with the antibody, as for panel a. Cells expressing hCD81 increased annexin V binding, while those expressing chimera VII(LEL) failed to do so, although 2F3 cells express normal levels of CD19. (d) 1C8/hCD9 or 1C8/II(N′-SEL) cells (see Fig. S5c in the supplemental material) were incubated with the biotinylated anti-hCD9 (50H.19) MAb or an isotype control MAb and cross-linked by streptavidin. Flow cytometry showed no increase annexin V binding. In histograms showing annexin V, specific MAbs are presented by solid lines and isotype controls by dotted lines. These data represent three independent experiments.
FIG. 7.
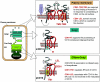
Distinct structural domains of CD81 function in different cellular compartments in B cells. The association between the CD81 LEL and CD19 (symbolized by a black ring) is maintained throughout maturation and trafficking of the molecules to the cell surface, where it is required for the activation of both molecules. However, the association via the CD81 LEL is not sufficient to support surface expression of CD19. ER exit and cell surface expression is supported by CD81 TM1 (highlighted by a green square). ER-retained molecules are sensitive to endo-H digestion of their high-mannose core oligosaccharide (symbolized by -○). In the Golgi, the CD81 N′ tail (highlighted by a blue square) is required for proper N glycosylation of CD19 (complexed oligosaccharides are indicated by Ψ). At the cell surface, engagement of CD81 with biotinylated (blue circle) anti-CD81 MAb cross-linked by streptavidin (SA) stimulates externalization of phosphatidylserine, increasing annexin V binding (purple). This membrane reorganization (curved arrow) requires CD81 TM2-TM3 (highlighted by a brown square) only in the context of the CD81 LEL.
Similar articles
- The tetraspanin CD81 is necessary for partitioning of coligated CD19/CD21-B cell antigen receptor complexes into signaling-active lipid rafts.
Cherukuri A, Shoham T, Sohn HW, Levy S, Brooks S, Carter R, Pierce SK. Cherukuri A, et al. J Immunol. 2004 Jan 1;172(1):370-80. doi: 10.4049/jimmunol.172.1.370. J Immunol. 2004. PMID: 14688345 - The tetraspanin CD81 regulates the expression of CD19 during B cell development in a postendoplasmic reticulum compartment.
Shoham T, Rajapaksa R, Boucheix C, Rubinstein E, Poe JC, Tedder TF, Levy S. Shoham T, et al. J Immunol. 2003 Oct 15;171(8):4062-72. doi: 10.4049/jimmunol.171.8.4062. J Immunol. 2003. PMID: 14530327 - CD19 is linked to the integrin-associated tetraspans CD9, CD81, and CD82.
Horváth G, Serru V, Clay D, Billard M, Boucheix C, Rubinstein E. Horváth G, et al. J Biol Chem. 1998 Nov 13;273(46):30537-43. doi: 10.1074/jbc.273.46.30537. J Biol Chem. 1998. PMID: 9804823 - Function of the tetraspanin molecule CD81 in B and T cells.
Levy S. Levy S. Immunol Res. 2014 May;58(2-3):179-85. doi: 10.1007/s12026-014-8490-7. Immunol Res. 2014. PMID: 24522698 Review. - Protein-protein interactions in the tetraspanin web.
Levy S, Shoham T. Levy S, et al. Physiology (Bethesda). 2005 Aug;20:218-24. doi: 10.1152/physiol.00015.2005. Physiology (Bethesda). 2005. PMID: 16024509 Review.
Cited by
- Human tetraspanin CD81 facilitates invasion of Salmonella enterica into human epithelial cells.
Alvarez KG, Goral L, Suwandi A, Lasswitz L, Zapatero-Belinchón FJ, Ehrhardt K, Nagarathinam K, Künnemann K, Krey T, Wiedemann A, Gerold G, Grassl GA. Alvarez KG, et al. Virulence. 2024 Dec;15(1):2399792. doi: 10.1080/21505594.2024.2399792. Epub 2024 Sep 24. Virulence. 2024. PMID: 39239914 Free PMC article. - Primary cilia-mediated regulation of microglial secretion in Alzheimer's disease.
Yeo S, Jang J, Jung HJ, Lee H, Choe Y. Yeo S, et al. Front Mol Biosci. 2023 Oct 23;10:1250335. doi: 10.3389/fmolb.2023.1250335. eCollection 2023. Front Mol Biosci. 2023. PMID: 37942288 Free PMC article. - The molecular mechanism of CD81 antibody inhibition of metastasis.
Abu-Saleh N, Kuo CC, Jiang W, Levy R, Levy S. Abu-Saleh N, et al. Proc Natl Acad Sci U S A. 2023 Jun 27;120(26):e2305042120. doi: 10.1073/pnas.2305042120. Epub 2023 Jun 20. Proc Natl Acad Sci U S A. 2023. PMID: 37339209 Free PMC article. - Understanding the Bioactivity and Prognostic Implication of Commonly Used Surface Antigens in Multiple Myeloma.
Lebel E, Nachmias B, Pick M, Gross Even-Zohar N, Gatt ME. Lebel E, et al. J Clin Med. 2022 Mar 25;11(7):1809. doi: 10.3390/jcm11071809. J Clin Med. 2022. PMID: 35407416 Free PMC article. Review. - Cancer cell-intrinsic resistance to BiTE therapy is mediated by loss of CD58 costimulation and modulation of the extrinsic apoptotic pathway.
Shen Y, Eng JS, Fajardo F, Liang L, Li C, Collins P, Tedesco D, Nolan-Stevaux O. Shen Y, et al. J Immunother Cancer. 2022 Mar;10(3):e004348. doi: 10.1136/jitc-2021-004348. J Immunother Cancer. 2022. PMID: 35296559 Free PMC article.
References
- Bradbury, L. E., V. S. Goldmacher, and T. F. Tedder. 1993. The CD19 signal transduction complex of B lymphocytes. Deletion of the CD19 cytoplasmic domain alters signal transduction but not complex formation with TAPA-1 and Leu 13. J. Immunol. 151:2915-2927. - PubMed
- Bradbury, L. E., G. S. Kansas, S. Levy, R. L. Evans, and T. F. Tedder. 1992. The CD19/CD21 signal transducing complex of human B lymphocytes includes the target of antiproliferative antibody-1 and Leu-13 molecules. J. Immunol. 149:2841-2850. - PubMed
- Brodsky, J. L., and A. A. McCracken. 1999. ER protein quality control and proteasome-mediated protein degradation. Semin. Cell Dev. Biol. 10:507-513. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases