Use of electrochemical DNA biosensors for rapid molecular identification of uropathogens in clinical urine specimens - PubMed (original) (raw)
doi: 10.1128/JCM.44.2.561-570.2006.
Mitra Mastali, Vincent Gau, Marc A Suchard, Annette K Møller, David A Bruckner, Jane T Babbitt, Yang Li, Jeffrey Gornbein, Elliot M Landaw, Edward R B McCabe, Bernard M Churchill, David A Haake
Affiliations
- PMID: 16455913
- PMCID: PMC1392664
- DOI: 10.1128/JCM.44.2.561-570.2006
Use of electrochemical DNA biosensors for rapid molecular identification of uropathogens in clinical urine specimens
Joseph C Liao et al. J Clin Microbiol. 2006 Feb.
Abstract
We describe the first species-specific detection of bacterial pathogens in human clinical fluid samples using a microfabricated electrochemical sensor array. Each of the 16 sensors in the array consisted of three single-layer gold electrodes-working, reference, and auxiliary. Each of the working electrodes contained one representative from a library of capture probes, each specific for a clinically relevant bacterial urinary pathogen. The library included probes for Escherichia coli, Proteus mirabilis, Pseudomonas aeruginosa, Enterocococcus spp., and the Klebsiella-Enterobacter group. A bacterial 16S rRNA target derived from single-step bacterial lysis was hybridized both to the biotin-modified capture probe on the sensor surface and to a second, fluorescein-modified detector probe. Detection of the target-probe hybrids was achieved through binding of a horseradish peroxidase (HRP)-conjugated anti-fluorescein antibody to the detector probe. Amperometric measurement of the catalyzed HRP reaction was obtained at a fixed potential of -200 mV between the working and reference electrodes. Species-specific detection of as few as 2,600 uropathogenic bacteria in culture, inoculated urine, and clinical urine samples was achieved within 45 min from the beginning of sample processing. In a feasibility study of this amperometric detection system using blinded clinical urine specimens, the sensor array had 100% sensitivity for direct detection of gram-negative bacteria without nucleic acid purification or amplification. Identification was demonstrated for 98% of gram-negative bacteria for which species-specific probes were available. When combined with a microfluidics-based sample preparation module, the integrated system could serve as a point-of-care device for rapid diagnosis of urinary tract infections.
Figures
FIG. 1.
Components and performance of the electrochemical sensor. (A) The 16-sensor array (2.5 by 7.5 cm) was microfabricated with a thin, optical-grade layer of gold electrodes deposited on plastic. Each sensor in the array contained three electrodes: a central working electrode, a circumferential reference electrode, and a short auxiliary electrode. (B) The chip mounter with contact pins for simultaneous reading of the current output from each of the sensors in the array. (C) Detection strategy. (1) Bacterial lysis to release 16S rRNA target (black dashed line). (2) Hybridization of the target with the fluorescein (green circle)-labeled detector probe (blue line). (3) Hybridization of the target with the biotin (red circle)-labeled capture probe (orange line). (4) Binding of anti-fluorescein antibody conjugated with HRP to the target-probe sandwich. (5) Generation of current by transfer of electrons to the electron transfer mediator, TMB. (D) Current output in an experiment involving a clinical urine specimen containing K. pneumoniae showing signal stabilization from all 16 sensors in the array within 60 seconds. Probe results were obtained by averaging the log10 current outputs from duplicate sensor readings at 60 seconds.
FIG. 2.
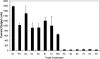
Specificities of _Enterobacteriaceae_-specific probe pairs. Positive signals were seen for all Enterobacteriaceae species tested but not for gram-positive uropathogens (Eo, Ef, Ss, and Sa) or P. aeruginosa (Pa). (See the footnote to Table 2 for bacterial species abbreviations.) Means and standard deviations of experiments performed in duplicate are shown. NC refers to the negative control experiments performed with capture and detector probes but without bacterial lysate.
FIG. 3.
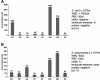
Direct, species-specific detection of uropathogens in representative clinical urine specimens using the electrochemical sensor array. Current output for each of the probe pairs in the array are shown in nanoamperes. The mean current output of duplicate sensors is shown above each bar; the error bars represent the standard deviations. The probe pair designation is shown below each bar, and the species specificity is given in Table 2. The urinalysis and microbiological characteristics of each specimen are shown to the right of the bar graph. The background signal level was determined by averaging the log10 results of the NC sensors and the sensors with the four lowest species-specific probe pairs (from among EC, PM, KE, PA, and EF). As described in the text, significant signals were 0.30 log unit (5 standard deviations) above background. (A) E. coli in this clinical urine specimen produced significant signals in the UNI, EB, and EC sensors despite high numbers of white blood cells (WBC). RBC, red blood cells. (B) 16S rRNA from as few as 4 × 104 K. pneumoniae cells/ml in urine produced significant signal levels in the UNI, EB, and KE sensors.
FIG. 4.
“UTI Chip” signal interpretation algorithm. The three-step algorithm used to interpret the results of electrochemical-sensor experiments on 78 specimens that met inclusion criteria is shown. Positive signals were those with a mean log of greater than 0.30 log unit (5 standard deviations) over background. “UNI” and “EB” are eubacterial- and _Enterobacteriaceae_-specific probe signals, respectively. “Bkgnd” is the background signal, as defined in the text. “MaxSpSp” refers to the maximum species-specific signal. Clinical microbiology results are given in shaded boxes. Two-letter species abbreviations are given in the footnote of Table 2. NSG indicates “no significant growth.” NG indicates “no growth.” An asterisk indicates false-positive results, and false-negative results are underlined.
Similar articles
- Development of an advanced electrochemical DNA biosensor for bacterial pathogen detection.
Liao JC, Mastali M, Li Y, Gau V, Suchard MA, Babbitt J, Gornbein J, Landaw EM, McCabe ER, Churchill BM, Haake DA. Liao JC, et al. J Mol Diagn. 2007 Apr;9(2):158-68. doi: 10.2353/jmoldx.2007.060052. J Mol Diagn. 2007. PMID: 17384207 Free PMC article. - Optimal probe length and target location for electrochemical detection of selected uropathogens at ambient temperature.
Mastali M, Babbitt JT, Li Y, Landaw EM, Gau V, Churchill BM, Haake DA. Mastali M, et al. J Clin Microbiol. 2008 Aug;46(8):2707-16. doi: 10.1128/JCM.00423-08. Epub 2008 Jun 18. J Clin Microbiol. 2008. PMID: 18562584 Free PMC article. - Rapid, species-specific detection of uropathogen 16S rDNA and rRNA at ambient temperature by dot-blot hybridization and an electrochemical sensor array.
Sun CP, Liao JC, Zhang YH, Gau V, Mastali M, Babbitt JT, Grundfest WS, Churchill BM, McCabe ER, Haake DA. Sun CP, et al. Mol Genet Metab. 2005 Jan;84(1):90-9. doi: 10.1016/j.ymgme.2004.11.006. Epub 2004 Dec 13. Mol Genet Metab. 2005. PMID: 15639199 - Recent advances in biosensor based diagnosis of urinary tract infection.
Kumar MS, Ghosh S, Nayak S, Das AP. Kumar MS, et al. Biosens Bioelectron. 2016 Jun 15;80:497-510. doi: 10.1016/j.bios.2016.02.023. Epub 2016 Feb 9. Biosens Bioelectron. 2016. PMID: 26890825 Review. - Biosensing platforms for Vibrio bacteria detection based on whole cell and nucleic acid analysis: A review.
Da-Silva E, Baudart J, Barthelmebs L. Da-Silva E, et al. Talanta. 2018 Dec 1;190:410-422. doi: 10.1016/j.talanta.2018.07.092. Epub 2018 Aug 4. Talanta. 2018. PMID: 30172527 Review.
Cited by
- A Multiplex Electrochemical Biosensor for Bloodstream Infection Diagnosis.
Gao J, Jeffries L, Mach KE, Craft DW, Thomas NJ, Gau V, Liao JC, Wong PK. Gao J, et al. SLAS Technol. 2017 Aug;22(4):466-474. doi: 10.1177/2211068216651232. Epub 2016 May 25. SLAS Technol. 2017. PMID: 27226118 Free PMC article. - New trends in impedimetric biosensors for the detection of foodborne pathogenic bacteria.
Wang Y, Ye Z, Ying Y. Wang Y, et al. Sensors (Basel). 2012;12(3):3449-71. doi: 10.3390/s120303449. Epub 2012 Mar 12. Sensors (Basel). 2012. PMID: 22737018 Free PMC article. Review. - PCR-Independent Detection of Bacterial Species-Specific 16S rRNA at 10 fM by a Pore-Blockage Sensor.
Esfandiari L, Wang S, Wang S, Banda A, Lorenzini M, Kocharyan G, Monbouquette HG, Schmidt JJ. Esfandiari L, et al. Biosensors (Basel). 2016 Jul 22;6(3):37. doi: 10.3390/bios6030037. Biosensors (Basel). 2016. PMID: 27455337 Free PMC article. - Antimicrobial susceptibility testing using high surface-to-volume ratio microchannels.
Chen CH, Lu Y, Sin ML, Mach KE, Zhang DD, Gau V, Liao JC, Wong PK. Chen CH, et al. Anal Chem. 2010 Feb 1;82(3):1012-9. doi: 10.1021/ac9022764. Anal Chem. 2010. PMID: 20055494 Free PMC article. - Multiplexed DNA-modified electrodes.
Slinker JD, Muren NB, Gorodetsky AA, Barton JK. Slinker JD, et al. J Am Chem Soc. 2010 Mar 3;132(8):2769-74. doi: 10.1021/ja909915m. J Am Chem Soc. 2010. PMID: 20131780 Free PMC article.
References
- Abdel-Hamid, I., D. Ivnitski, P. Atanasov, and E. Wilkins. 1999. Flow-through immunofiltration assay system for rapid detection of E. coli O157:H7. Biosens. Bioelectron. 14:309-316. - PubMed
- Albers, J., T. Grunwald, E. Nebling, G. Piechotta, and R. Hintsche. 2003. Electrical biochip technology—a tool for microarrays and continuous monitoring. Anal. Bioanal. Chem. 377:521-527. - PubMed
- Bard, A. J., and L. R. Faulkner. 2001. Electrochemical methods fundamentals and applications, 2nd ed. John Wiley & Sons, Inc., Hoboken, N.J.
- Basu, M., S. Seggerson, J. Henshaw, J. Jiang, R. del A Cordona, C. Lefave, P. J. Boyle, A. Miller, M. Pugia, and S. Basu. 2004. Nano-biosensor development for bacterial detection during human kidney infection: use of glycoconjugate-specific antibody-bound gold NanoWire arrays (GNWA). Glycoconj. J. 21:487-496. - PubMed
- Campbell, C. N., D. Gal, N. Cristler, C. Banditrat, and A. Heller. 2002. Enzyme-amplified amperometric sandwich test for RNA and DNA. Anal. Chem. 74:158-162. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous