Dynamic transition of a methanogenic population in response to the concentration of volatile fatty acids in a thermophilic anaerobic digester - PubMed (original) (raw)
Dynamic transition of a methanogenic population in response to the concentration of volatile fatty acids in a thermophilic anaerobic digester
Tomoyuki Hori et al. Appl Environ Microbiol. 2006 Feb.
Abstract
In this study, the microbial community succession in a thermophilic methanogenic bioreactor under deteriorative and stable conditions that were induced by acidification and neutralization, respectively, was investigated using PCR-mediated single-strand conformation polymorphism (SSCP) based on the 16S rRNA gene, quantitative PCR, and fluorescence in situ hybridization (FISH). The SSCP analysis indicated that the archaeal community structure was closely correlated with the volatile fatty acid (VFA) concentration, while the bacterial population was impacted by pH. The archaeal community consisted mainly of two species of hydrogenotrophic methanogen (i.e., a Methanoculleus sp. and a Methanothermobacter sp.) and one species of aceticlastic methanogen (i.e., a Methanosarcina sp.). The quantitative PCR of the 16S rRNA gene from each methanogen revealed that the Methanoculleus sp. predominated among the methanogens during operation under stable conditions in the absence of VFAs. Accumulation of VFAs induced a dynamic transition of hydrogenotrophic methanogens, and in particular, a drastic change (i.e., an approximately 10,000-fold increase) in the amount of the 16S rRNA gene from the Methanothermobacter sp. The predominance of the one species of hydrogenotrophic methanogen was replaced by that of the other in response to the VFA concentration, suggesting that the dissolved hydrogen concentration played a decisive role in the predominance. The hydrogenotrophic methanogens existed close to bacteria in aggregates, and a transition of the associated bacteria was also observed by FISH analyses. The degradation of acetate accumulated during operation under deteriorative conditions was concomitant with the selective proliferation of the Methanosarcina sp., indicating effective acetate degradation by the aceticlastic methanogen. The simple methanogenic population in the thermophilic anaerobic digester significantly responded to the environmental conditions, especially to the concentration of VFAs.
Figures
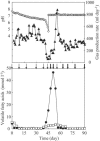
FIG. 1.
Change in reactor performance of the thermophilic methanogenic process: ⋄, pH; ▴, gas production rate; •, acetate concentration; □, propionate concentration. Arrows indicate the sampling points for SSCP analysis and quantitative PCR. Arrows in boldface indicate the sampling points for FISH.
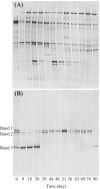
FIG. 2.
Microbial community structure in the thermophilic methanogenic process as determined using PCR-SSCP analysis. (A) Bacterial fingerprint; (B) archaeal fingerprint. Bands 1, 2, and 3 in panel B were closely related to Methanothermobacter thermautotrophicus (accession number AE000940; 100% sequence similarity), Methanosarcina thermophila (M59140; 98% sequence similarity), and Methanoculleus thermophilicus (AB065297; 100% sequence similarity), respectively.
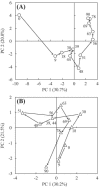
FIG. 3.
Principal-component analysis of the SSCP data. (A) PCA plot for the bacterial fingerprint; (B) PCA plot for the archaeal fingerprint. The successive time points (days) are connected by arrows and indicated by numbers.
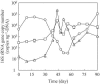
FIG. 4.
Transition of the 16S rRNA gene copy numbers of the methanogens Methanoculleus sp. (○), Methanosarcina sp. (□), and _Methanothermobacte_r sp. (▵). Two trials were conducted to analyze each sample. The variations between the duplications were less than 1% of the mean.
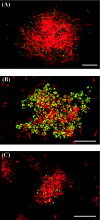
FIG. 5.
Fluorescence in situ hybridization of methanogenic microflora viewed by confocal laser scanning microscopy. (A) Day 48; (B) day 63; (C) day 90. The methanogenic microflora was simultaneously hybridized with Alexa488-labeled archaeal probe ARC915 (green) and rhodamine-labeled bacterial probe EUB338 (red). Representative fields of vision are shown. Bars, 20 μm.
Similar articles
- Acetoclastic and hydrogenotrophic methane production and methanogenic populations in an acidic West-Siberian peat bog.
Kotsyurbenko OR, Chin KJ, Glagolev MV, Stubner S, Simankova MV, Nozhevnikova AN, Conrad R. Kotsyurbenko OR, et al. Environ Microbiol. 2004 Nov;6(11):1159-73. doi: 10.1111/j.1462-2920.2004.00634.x. Environ Microbiol. 2004. PMID: 15479249 - Analysis of microbial community structure and diversity in the thermophilic anaerobic digestion of waste activated sludge.
Kobayashi T, Li YY, Harada H. Kobayashi T, et al. Water Sci Technol. 2008;57(8):1199-205. doi: 10.2166/wst.2008.079. Water Sci Technol. 2008. PMID: 18469390 - Inhibitory effects of ferrihydrite on a thermophilic methanogenic community.
Yamada C, Kato S, Ueno Y, Ishii M, Igarashi Y. Yamada C, et al. Microbes Environ. 2014;29(2):227-30. doi: 10.1264/jsme2.me14026. Epub 2014 May 23. Microbes Environ. 2014. PMID: 24859310 Free PMC article. - Obstacles faced by methanogenic archaea originating from substrate-driven toxicants in anaerobic digestion.
Cai Y, Zheng Z, Wang X. Cai Y, et al. J Hazard Mater. 2021 Feb 5;403:123938. doi: 10.1016/j.jhazmat.2020.123938. Epub 2020 Sep 14. J Hazard Mater. 2021. PMID: 33264986 Review. - Syntrophic associations in methanogenic degradation.
Schink B. Schink B. Prog Mol Subcell Biol. 2006;41:1-19. doi: 10.1007/3-540-28221-1_1. Prog Mol Subcell Biol. 2006. PMID: 16623386 Review. No abstract available.
Cited by
- Growth and Break-Up of Methanogenic Granules Suggests Mechanisms for Biofilm and Community Development.
Trego AC, Galvin E, Sweeney C, Dunning S, Murphy C, Mills S, Nzeteu C, Quince C, Connelly S, Ijaz UZ, Collins G. Trego AC, et al. Front Microbiol. 2020 Jun 3;11:1126. doi: 10.3389/fmicb.2020.01126. eCollection 2020. Front Microbiol. 2020. PMID: 32582085 Free PMC article. - Methanosarcina Play an Important Role in Anaerobic Co-Digestion of the Seaweed Ulva lactuca: Taxonomy and Predicted Metabolism of Functional Microbial Communities.
FitzGerald JA, Allen E, Wall DM, Jackson SA, Murphy JD, Dobson AD. FitzGerald JA, et al. PLoS One. 2015 Nov 10;10(11):e0142603. doi: 10.1371/journal.pone.0142603. eCollection 2015. PLoS One. 2015. PMID: 26555136 Free PMC article. - Relating Anaerobic Digestion Microbial Community and Process Function.
Venkiteshwaran K, Bocher B, Maki J, Zitomer D. Venkiteshwaran K, et al. Microbiol Insights. 2016 Apr 20;8(Suppl 2):37-44. doi: 10.4137/MBI.S33593. eCollection 2015. Microbiol Insights. 2016. PMID: 27127410 Free PMC article. Review. - Effect of biowaste sludge maturation on the diversity of thermophilic bacteria and archaea in an anaerobic reactor.
Goberna M, Insam H, Franke-Whittle IH. Goberna M, et al. Appl Environ Microbiol. 2009 Apr;75(8):2566-72. doi: 10.1128/AEM.02260-08. Epub 2009 Feb 13. Appl Environ Microbiol. 2009. PMID: 19218417 Free PMC article.
References
- Ahring, B. K. 1995. Methanogenesis in thermophilic biogas reactors. Antonie Leeuwenhoek 67:91-102. - PubMed
- Ahring, B. K., M. Sandberg, and I. Angelidaki. 1995. Volatile fatty acids as indicators of process imbalance in anaerobic digestors. Appl. Microbiol. Biotechnol. 41:559-565.
- Bjornsson, L., M. Murto, T. G. Jantsch, and B. Mattiasson. 2001. Evaluation of new methods for the monitoring of alkalinity, dissolved hydrogen and the microbial community in anaerobic digestion. Water Res. 35:2833-2840. - PubMed
- Boone, D. R., and R. W. Castenholz. 2001. The Archaea and the deeply branching and phototrophic Bacteria. In G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, N.Y.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases