Simple fold composition and modular architecture of the nuclear pore complex - PubMed (original) (raw)
Simple fold composition and modular architecture of the nuclear pore complex
Damien Devos et al. Proc Natl Acad Sci U S A. 2006.
Abstract
The nuclear pore complex (NPC) consists of multiple copies of approximately 30 different proteins [nucleoporins (nups)], forming a channel in the nuclear envelope that mediates macromolecular transport between the cytosol and the nucleus. With <5% of the nup residues currently available in experimentally determined structures, little is known about the detailed structure of the NPC. Here, we use a combined computational and biochemical approach to assign folds for approximately 95% of the residues in the yeast and vertebrate nups. These fold assignments suggest an underlying simplicity in the composition and modularity in the architecture of all eukaryotic NPCs. The simplicity in NPC composition is reflected in the presence of only eight fold types, with the three most frequent folds accounting for approximately 85% of the residues. The modularity in NPC architecture is reflected in its hierarchical and symmetrical organization that partitions the predicted nup folds into three groups: the transmembrane group containing transmembrane helices and a cadherin fold, the central scaffold group containing beta-propeller and alpha-solenoid folds, and the peripheral FG group containing predominantly the FG repeats and the coiled-coil fold. Moreover, similarities between structures in coated vesicles and those in the NPC support our prior hypothesis for their common evolutionary origin in a progenitor protocoatomer. The small number of predicted fold types in the NPC and their internal symmetries suggest that the bulk of the NPC structure has evolved through extensive motif and gene duplication from a simple precursor set of only a few proteins.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
Fig. 1.
Predicted secondary structure and fold types of the nups. The names of the nups are boxed according to the group they define: transmembrane (pink), scaffold (orange), and FG repeat (green). The black horizontal lines on the right represent the sequence of each yeast nup. The predicted α-helices (magenta) and β-strands (cyan) are indicated by bars above each line. The height of the bars is proportional to the confidence of the prediction (39). Predicted transmembrane helices are shown in green, coiled coils are shown in red, FG repeats are shown in black, and unstructured regions are shown by an empty box. An orange block underlines regions of >50 residues to which a fold type could not be assigned. Representative models of the nup domains are colored according to the fold type and are shown on the left. Models are not to scale for visualization reasons. There are eight fold types. First, a TMH segment (green) is a hydrophobic 15- to 30-residue helical segment that spans the membrane. Second, cadherin domains (dark blue) have ≈110 residues that fold into a seven-stranded β-sandwich structure. Third, β-propellers (cyan) contain several blades arranged radially around a central axis, each blade consisting of a four-stranded antiparallel β-sheet. Fourth, α-solenoid domains are composed of numerous pairs of antiparallel α-helices stacked to form a solenoid. Fifth, coiled coils (red) generally display seven-residue repeats where the first and fourth residues of an α-helix are often hydrophobic. The coiled-coil structure is formed by helices (generally two) twisting together to bury their hydrophobic seams. Sixth, disordered FG-repeat segments are indicated schematically by a black curve. Seventh, the autoproteolytic domain of Nup98 (yellow) adopts a half-open, β-sandwich-like fold dominated by a large β-sheet with helices capping two of its ends. Finally, the RRM (orange) is a two-layer α/β sandwich typically found in proteins involved in RNA binding.
Fig. 2.
Mapping of domains of the yeast nups by protease accessibility laddering. The gels show immunoblots of limited proteolysis digests for Protein A-tagged versions of the nups. Each full-length nup is indicated by an arrow on the left side of the gel, as are the sizes of marker proteins (expressed in kDa). Secondary structure predictions are shown by using the conventions in Fig. 1. Proteolytic cleavage sites are identified by small, medium, and large arrows for weak, medium, and strong susceptibility sites, respectively. Where necessary, uncertainties in the precise cleavage positions are indicated by lines to the left of the sequence.
Fig. 3.
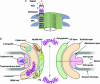
Simplicity of the fold composition and modular architecture of the NPC. (a) The schematic structure and hierarchy of the NPC. Most of the nups consist of sequence repeats (step I). The nups assemble into multiple copies to form each half-spoke (step II) that dimerize to form the spokes (step III), which are themselves repeated eight times to form the complete NPC (step IV). (b) The architecture of the NPC ring, viewed in the plane of the NE, is segregated into the membrane (pink), scaffold (orange), and FG (green) groups. The domain fold types assigned to each group are indicated on the left side of the schema. The schema illustrates the coarse organization of the NPC and is not a precise map of the three-dimensional nup locations.
Similar articles
- Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor.
DeGrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, Rout MP, Chait BT. DeGrasse JA, et al. Mol Cell Proteomics. 2009 Sep;8(9):2119-30. doi: 10.1074/mcp.M900038-MCP200. Epub 2009 Jun 13. Mol Cell Proteomics. 2009. PMID: 19525551 Free PMC article. - Natively Unfolded FG Repeats Stabilize the Structure of the Nuclear Pore Complex.
Onischenko E, Tang JH, Andersen KR, Knockenhauer KE, Vallotton P, Derrer CP, Kralt A, Mugler CF, Chan LY, Schwartz TU, Weis K. Onischenko E, et al. Cell. 2017 Nov 2;171(4):904-917.e19. doi: 10.1016/j.cell.2017.09.033. Epub 2017 Oct 12. Cell. 2017. PMID: 29033133 Free PMC article. - Nucleoporin's Like Charge Regions Are Major Regulators of FG Coverage and Dynamics Inside the Nuclear Pore Complex.
Peyro M, Soheilypour M, Ghavami A, Mofrad MR. Peyro M, et al. PLoS One. 2015 Dec 11;10(12):e0143745. doi: 10.1371/journal.pone.0143745. eCollection 2015. PLoS One. 2015. PMID: 26658558 Free PMC article. - [Nuclear pores: from yeast to higher eukaryotes].
Doye V. Doye V. J Soc Biol. 2002;196(4):349-54. J Soc Biol. 2002. PMID: 12645306 Review. French. - Membrane-coating lattice scaffolds in the nuclear pore and vesicle coats: commonalities, differences, challenges.
Leksa NC, Schwartz TU. Leksa NC, et al. Nucleus. 2010 Jul-Aug;1(4):314-8. doi: 10.4161/nucl.1.4.11798. Epub 2010 Mar 12. Nucleus. 2010. PMID: 21327078 Free PMC article. Review.
Cited by
- A role for the inositol kinase Ipk1 in ciliary beating and length maintenance.
Sarmah B, Winfrey VP, Olson GE, Appel B, Wente SR. Sarmah B, et al. Proc Natl Acad Sci U S A. 2007 Dec 11;104(50):19843-8. doi: 10.1073/pnas.0706934104. Epub 2007 Dec 3. Proc Natl Acad Sci U S A. 2007. PMID: 18056639 Free PMC article. - Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport.
Terry LJ, Wente SR. Terry LJ, et al. Eukaryot Cell. 2009 Dec;8(12):1814-27. doi: 10.1128/EC.00225-09. Epub 2009 Oct 2. Eukaryot Cell. 2009. PMID: 19801417 Free PMC article. Review. - Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Akey CW, Singh D, Ouch C, Echeverria I, Nudelman I, Varberg JM, Yu Z, Fang F, Shi Y, Wang J, Salzberg D, Song K, Xu C, Gumbart JC, Suslov S, Unruh J, Jaspersen SL, Chait BT, Sali A, Fernandez-Martinez J, Ludtke SJ, Villa E, Rout MP. Akey CW, et al. Cell. 2022 Jan 20;185(2):361-378.e25. doi: 10.1016/j.cell.2021.12.015. Epub 2022 Jan 3. Cell. 2022. PMID: 34982960 Free PMC article. - Nuclear pore complex biogenesis.
Fernandez-Martinez J, Rout MP. Fernandez-Martinez J, et al. Curr Opin Cell Biol. 2009 Aug;21(4):603-12. doi: 10.1016/j.ceb.2009.05.001. Epub 2009 Jun 11. Curr Opin Cell Biol. 2009. PMID: 19524430 Free PMC article. Review. - Enriching the pore: splendid complexity from humble origins.
Field MC, Koreny L, Rout MP. Field MC, et al. Traffic. 2014 Feb;15(2):141-56. doi: 10.1111/tra.12141. Epub 2014 Jan 8. Traffic. 2014. PMID: 24279500 Free PMC article. Review.
References
- Rout M. P., Aitchison J. D. J. Biol. Chem. 2001;276:16593–16596. - PubMed
- Suntharalingam M., Wente S. R. Dev. Cell. 2003;4:775–789. - PubMed
- Beck M., Forster F., Ecke M., Plitzko J. M., Melchior F., Gerisch G., Baumeister W., Medalia O. Science. 2004;306:1387–1390. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM62529/GM/NIGMS NIH HHS/United States
- R33 CA089810/CA/NCI NIH HHS/United States
- RR022220/RR/NCRR NIH HHS/United States
- P41 RR000862/RR/NCRR NIH HHS/United States
- U54 RR022220/RR/NCRR NIH HHS/United States
- CA89810/CA/NCI NIH HHS/United States
- P50 GM062529/GM/NIGMS NIH HHS/United States
- R01 GM054762/GM/NIGMS NIH HHS/United States
- R29 GM054762/GM/NIGMS NIH HHS/United States
- GM54762/GM/NIGMS NIH HHS/United States
- GM062427/GM/NIGMS NIH HHS/United States
- RR00862/RR/NCRR NIH HHS/United States
- R01 GM062427/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases