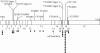Mucolipidosis II (I-cell disease) and mucolipidosis IIIA (classical pseudo-hurler polydystrophy) are caused by mutations in the GlcNAc-phosphotransferase alpha / beta -subunits precursor gene - PubMed (original) (raw)
Mucolipidosis II (I-cell disease) and mucolipidosis IIIA (classical pseudo-hurler polydystrophy) are caused by mutations in the GlcNAc-phosphotransferase alpha / beta -subunits precursor gene
Mariko Kudo et al. Am J Hum Genet. 2006 Mar.
Abstract
Mucolipidosis II (MLII; I-cell disease) and mucolipidosis IIIA (MLIIIA; classical pseudo-Hurler polydystrophy) are diseases in which the activity of the uridine diphosphate (UDP)-N-acetylglucosamine:lysosomal enzyme N-acetylglucosamine-1-phosphotransferase (GlcNAc-phosphotransferase) is absent or reduced, respectively. In the absence of mannose phosphorylation, trafficking of lysosomal hydrolases to the lysosome is impaired. In these diseases, mistargeted lysosomal hydrolases are secreted into the blood, resulting in lysosomal deficiency of many hydrolases and a storage-disease phenotype. To determine whether these diseases are caused by mutations in the GlcNAc-phosphotransferase alpha / beta -subunits precursor gene (GNPTAB), we sequenced GNPTAB exons and flanking intronic sequences and measured GlcNAc-phosphotransferase activity in patient fibroblasts. We identified 15 different mutations in GNPTAB from 18 pedigrees with MLII or MLIIIA and demonstrated that these two diseases are allelic. Mutations in both alleles were identified in each case, which demonstrated that GNPTAB mutations are the cause of both diseases. Some pedigrees had identical mutations. One frameshift mutation (truncation at amino acid 1171) predominated and was found in both MLII and MLIIIA. This mutation was found in combination with severe mutations (i.e., mutations preventing the generation of active enzyme) in MLII and with mild mutations (i.e., mutations allowing the generation of active enzyme) in MLIIIA. Some cases of MLII and MLIIIA were the result of mutations that cause aberrant splicing. Substitutions were inside the invariant splice-site sequence in MLII and were outside it in MLIIIA. When the mutations were analyzed along with GlcNAc-phosphotransferase activity, it was possible to confidently distinguish these two clinically related but distinct diseases. We propose criteria for distinguishing these two disorders by a combination of mutation detection and GlcNAc-phosphotransferase activity determination.
Figures
Figure 1
Location of mutations on the GlcNAc-phosphotransferase α/β–subunits precursor cDNA. The vertical lines indicate positions of mutations. Numbers below the boxes are exon numbers. Exons that are deleted as a result of splice-site mutations are shaded. An asterisk (*) before the name of a mutation indicates that the frameshift is caused by aberrant splicing. Blackened squares under the cDNA show the frequency of the mutant alleles in this study.
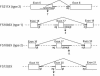
Figure 2
Structure of mutations causing aberrant splicing. Mutations and the resulting mRNA splicing that cause F211X (type 2), FS1085X (type 1), FS1085X (type 2), and FS1202X are shown. Nucleotide substitutions are indicated (arrows). RNA was analyzed by RT-PCR with oligo(dT) or random hexamer primers, and the amplified products were sequenced.
Figure 3
Comparison of frameshift mutations that cause termination at amino acid 211. Deletions in the cDNA sequence change the reading frame in FS211X (type 1) and FS211X (type 2). Nucleotides deleted from the cDNA are boxed. The wild-type reading frame is indicated by spacing; the abnormal reading frame, by brackets.
Similar articles
- Molecular analysis of the GNPTAB and GNPTG genes in 13 patients with mucolipidosis type II or type III - identification of eight novel mutations.
Encarnação M, Lacerda L, Costa R, Prata MJ, Coutinho MF, Ribeiro H, Lopes L, Pineda M, Ignatius J, Galvez H, Mustonen A, Vieira P, Lima MR, Alves S. Encarnação M, et al. Clin Genet. 2009 Jul;76(1):76-84. doi: 10.1111/j.1399-0004.2009.01185.x. Clin Genet. 2009. PMID: 19659762 - Analysis of mucolipidosis II/III GNPTAB missense mutations identifies domains of UDP-GlcNAc:lysosomal enzyme GlcNAc-1-phosphotransferase involved in catalytic function and lysosomal enzyme recognition.
Qian Y, van Meel E, Flanagan-Steet H, Yox A, Steet R, Kornfeld S. Qian Y, et al. J Biol Chem. 2015 Jan 30;290(5):3045-56. doi: 10.1074/jbc.M114.612507. Epub 2014 Dec 11. J Biol Chem. 2015. PMID: 25505245 Free PMC article. - GNPTAB missense mutations cause loss of GlcNAc-1-phosphotransferase activity in mucolipidosis type II through distinct mechanisms.
Ludwig NF, Velho RV, Sperb-Ludwig F, Acosta AX, Ribeiro EM, Kim CA, Gandelman Horovitz DD, Boy R, Rodovalho-Doriqui MJ, Lourenço CM, Santos ES, Braulke T, Pohl S, Schwartz IVD. Ludwig NF, et al. Int J Biochem Cell Biol. 2017 Nov;92:90-94. doi: 10.1016/j.biocel.2017.09.006. Epub 2017 Sep 14. Int J Biochem Cell Biol. 2017. PMID: 28918368 - Mannose phosphorylation in health and disease.
Kollmann K, Pohl S, Marschner K, Encarnação M, Sakwa I, Tiede S, Poorthuis BJ, Lübke T, Müller-Loennies S, Storch S, Braulke T. Kollmann K, et al. Eur J Cell Biol. 2010 Jan;89(1):117-23. doi: 10.1016/j.ejcb.2009.10.008. Epub 2009 Nov 28. Eur J Cell Biol. 2010. PMID: 19945768 Review. - Molecular analysis of the GlcNac-1-phosphotransferase.
Braulke T, Pohl S, Storch S. Braulke T, et al. J Inherit Metab Dis. 2008 Apr;31(2):253-7. doi: 10.1007/s10545-008-0862-5. Epub 2008 Apr 15. J Inherit Metab Dis. 2008. PMID: 18425436 Review.
Cited by
- Mucolipidosis III GNPTG Missense Mutations Cause Misfolding of the γ Subunit of GlcNAc-1-Phosphotransferase.
van Meel E, Kornfeld S. van Meel E, et al. Hum Mutat. 2016 Jul;37(7):623-6. doi: 10.1002/humu.22993. Epub 2016 Apr 22. Hum Mutat. 2016. PMID: 27038293 Free PMC article. - Stuttering as a spectrum disorder: A hypothesis.
SheikhBahaei S, Millwater M, Maguire GA. SheikhBahaei S, et al. Curr Res Neurobiol. 2023 Nov 1;5:100116. doi: 10.1016/j.crneur.2023.100116. eCollection 2023. Curr Res Neurobiol. 2023. PMID: 38020803 Free PMC article. Review. - Genetic approaches to understanding the causes of stuttering.
Drayna D, Kang C. Drayna D, et al. J Neurodev Disord. 2011 Dec;3(4):374-80. doi: 10.1007/s11689-011-9090-7. Epub 2011 Aug 18. J Neurodev Disord. 2011. PMID: 21850444 Free PMC article. - Deficiency in the endocytic adaptor proteins PHETA1/2 impairs renal and craniofacial development.
Ates KM, Wang T, Moreland T, Veeranan-Karmegam R, Ma M, Jeter C, Anand P, Wenzel W, Kim HG, Wolfe LA, Stephen J, Adams DR, Markello T, Tifft CJ, Settlage R, Gahl WA, Gonsalvez GB, Malicdan MC, Flanagan-Steet H, Pan YA. Ates KM, et al. Dis Model Mech. 2020 May 26;13(5):dmm041913. doi: 10.1242/dmm.041913. Dis Model Mech. 2020. PMID: 32152089 Free PMC article. - Emptying the stores: lysosomal diseases and therapeutic strategies.
Platt FM. Platt FM. Nat Rev Drug Discov. 2018 Feb;17(2):133-150. doi: 10.1038/nrd.2017.214. Epub 2017 Nov 17. Nat Rev Drug Discov. 2018. PMID: 29147032 Review.
References
Web Resources
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for BAC 14951 containing GNPTAB [accession number AC005409], cDNA encoding GlcNAc-phosphotransferase α/β–subunit precursor [accession number AY687932], and the following exons and the flanking intronic sequences: exon 1 [accession number BV677459], exon 2 [accession number BV677460], exons 3 and 4 [accession number BV677461], exon 5 [accession number BV677462], exons 6 and 7 [accession number BV677463], exons 8, 9, and 10 [accession number BV677464], exon 11 [accession number BV677465], exon 12 [accession number BV677466], exon 13 [accession number BV677467], exons 14 and 15 [accession number BV677468], exon 16 [accession number BV677469], exons 17 and 18 [accession number BV677470], exon 19 [accession number BV677471], exon 20 [accession number BV677472], and exon 21 [accession number BV677463])
- IUBMB (International Union of Biochemistry and Molecular Biology) Enzyme Nomenclature, http://www.chem.qmul.ac.uk/iubmb/enzyme/ (for accession numbers EC 2.7.8.17 and EC 3.1.4.45)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for MLII, MLIIIA, and MLIIIC)
References
- Canfield WM, Bao M, Pan J, D’Souza A, Brewer K, Pan H, Roe B, Raas-Rothschild A (1998) Mucolipidosis II and mucolipidosis IIIA are caused by mutations in the GlcNAc-phosphotransferase α/β gene on chromosome 12p. Am J Hum Genet Suppl 63:A15
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous