CLIP-170 homologue and NUDE play overlapping roles in NUDF localization in Aspergillus nidulans - PubMed (original) (raw)
CLIP-170 homologue and NUDE play overlapping roles in NUDF localization in Aspergillus nidulans
Vladimir P Efimov et al. Mol Biol Cell. 2006 Apr.
Abstract
Proteins in the cytoplasmic dynein pathway accumulate at the microtubule plus end, giving the appearance of comets when observed in live cells. The targeting mechanism for NUDF (LIS1/Pac1) of Aspergillus nidulans, a key component of the dynein pathway, has not been clear. Previous studies have demonstrated physical interactions of NUDF/LIS1/Pac1 with both NUDE/NUDEL/Ndl1 and CLIP-170/Bik1. Here, we have identified the A. nidulans CLIP-170 homologue, CLIPA. The clipA deletion did not cause an obvious nuclear distribution phenotype but affected cytoplasmic microtubules in an unexpected manner. Although more microtubules failed to undergo long-range growth toward the hyphal tip at 32 degrees C, those that reached the hyphal tip were less likely to undergo catastrophe. Thus, in addition to acting as a growth-promoting factor, CLIPA also promotes microtubule dynamics. In the absence of CLIPA, green fluorescent protein-labeled cytoplasmic dynein heavy chain, p150(Glued) dynactin, and NUDF were all seen as plus-end comets at 32 degrees C. However, under the same conditions, deletion of both clipA and nudE almost completely abolished NUDF comets, although nudE deletion itself did not cause a dramatic change in NUDF localization. Based on these results, we suggest that CLIPA and NUDE both recruit NUDF to the microtubule plus end. The plus-end localization of CLIPA itself seems to be regulated by different mechanisms under different physiological conditions. Although the KipA kinesin (Kip2/Tea2 homologue) did not affect plus-end localization of CLIPA at 32 degrees C, it was required for enhancing plus-end accumulation of CLIPA at an elevated temperature (42 degrees C).
Figures
Figure 1.
(A) Modular organization of the A. nidulans homologue of CLIP-170, CLIPA. Domains are drawn approximately to scale. (B) Alignment of zinc finger motifs from several CLIP-170 homologues. The regions shown are (top to bottom) human CLIP-170 (accession no. A43336) residues 1317-1356 (first zinc finger) and 1357-1392 (second zinc finger), A. nidulans CLIPA (AN1475.2) residues 537-578 (first zinc finger) and 635-668 (second zinc finger), S. pombe Tip1 (CAB16742) residues 422-461, and S. cerevisiae Bik1 (NP_009901) residues 400-440.
Figure 2.
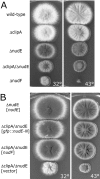
(A) Colonies of the Δ_clipA_ deletion mutant, wild type, Δ_nudE_, Δ_nudF_, and Δ_clipA_/Δ_nudE_ double deletion mutant. (B) The Δ_clipA_/Δ_nudE_ double deletion is suppressed by multiple copies of nudF and the N-terminal domain of nudE. The strains were transformed with the multicopy plasmid pAid carrying a gene indicated in square brackets. Δ_nudE_ strain transformed with pAid::nudE (top) serves as a wild-type control. Strains were grown at indicated temperature on YAG + UU plates for 3 d.
Figure 3.
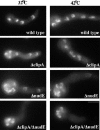
4,6-Diamidino-2-phenylindole staining of wild-type and mutant cells. The cells were grown on YG + UU medium at indicated temperature for 8 h. Bar, 5 μm.
Figure 4.
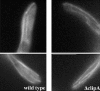
Cytoplasmic microtubules are able to extend to the hyphal tip in the Δ_clipA_ mutant. Cells were grown in minimal glycerol medium for overnight at 32°C. Bar, 5 μm.
Figure 5.
(A) GFP-NUDF comets are present in the Δ_clipA_ and Δ_nudE_ single deletion mutants but not in the Δ_clipA_/Δ_nudE_ double deletion mutant. GFP-NUDA comets are present in the double mutant grown under the same condition. Strains were grown in liquid minimal glycerol medium at 32°C for overnight before observation. Bar, 5 μm. (B) A Western blot showing that the protein level of GFP-NUDF did not change significantly in the double deletion mutant.
Figure 6.
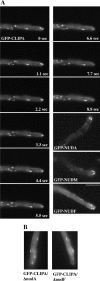
(A) Localization of GFP-CLIPA. GFP-NUDA, GFP-NUDM, and GFP-NUDF localization are shown for comparison. Cells were grown in minimal glycerol medium overnight at 32°C, and images were taken at 32°C. Images were taken every 1 s with 0.1-s exposure. (B) GFP-CLIPA comets in Δ_nudA_ and Δ_nudF_ strains. Bar, 5 μm.
Figure 7.
Observation of a nonfluorescent speckle followed by a trailing fluorescent dot (indicated by arrows) at the trailing end of a comet. The same images are shown in the bottom panel with two lines. The top line indicates that the leading end of the comet apparently moved forward. The bottom line shows that the position of the nonfluorescent spot did not change significantly. Images were taken at room temperature (at ∼25°C). Bar, 4 μm.
Figure 8.
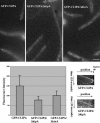
GFP-CLIPA localization in wild-type, Δ_kipA_, and Δ_kinA_ cells at 42°C. Cells were grown in minimal glycerol medium overnight at 42°C, and images were taken at 42°C. Bar, 5 μm. Bottom left, means and standard deviations of fluorescence intensity (arbitrary units) of comets are shown (wild type: n = 18; Δ_kipA_: n = 31; and Δ_kinA_: n = 21). Bottom right, kymographs showing the path of a comet in a wild-type cell (note that backtracking is obvious in the bottom half of the kymograph), and that of a comet in the Δ_kipA_ mutant moving toward the hyphal tip. The slopes in the two kymographs are similar, indicating that these two comets moved at about the same rates toward the hyphal tip. The kymograph showing the comet in Δ_kipA_ is less clear due to the lower fluorescence intensity of the comet.
Similar articles
- Accumulation of cytoplasmic dynein and dynactin at microtubule plus ends in Aspergillus nidulans is kinesin dependent.
Zhang J, Li S, Fischer R, Xiang X. Zhang J, et al. Mol Biol Cell. 2003 Apr;14(4):1479-88. doi: 10.1091/mbc.e02-08-0516. Mol Biol Cell. 2003. PMID: 12686603 Free PMC article. - The LIS1-related protein NUDF of Aspergillus nidulans and its interaction partner NUDE bind directly to specific subunits of dynein and dynactin and to alpha- and gamma-tubulin.
Hoffmann B, Zuo W, Liu A, Morris NR. Hoffmann B, et al. J Biol Chem. 2001 Oct 19;276(42):38877-84. doi: 10.1074/jbc.M106610200. Epub 2001 Aug 16. J Biol Chem. 2001. PMID: 11509576 Retracted. - The p25 subunit of the dynactin complex is required for dynein-early endosome interaction.
Zhang J, Yao X, Fischer L, Abenza JF, Peñalva MA, Xiang X. Zhang J, et al. J Cell Biol. 2011 Jun 27;193(7):1245-55. doi: 10.1083/jcb.201011022. J Cell Biol. 2011. PMID: 21708978 Free PMC article. - Microtubule plus ends, motors, and traffic of Golgi membranes.
Vaughan KT. Vaughan KT. Biochim Biophys Acta. 2005 Jul 10;1744(3):316-24. doi: 10.1016/j.bbamcr.2005.05.001. Biochim Biophys Acta. 2005. PMID: 15950296 Review. - CLIPs and CLASPs and cellular dynamics.
Galjart N. Galjart N. Nat Rev Mol Cell Biol. 2005 Jun;6(6):487-98. doi: 10.1038/nrm1664. Nat Rev Mol Cell Biol. 2005. PMID: 15928712 Review.
Cited by
- Dynamics of multiple nuclei in Ashbya gossypii hyphae depend on the control of cytoplasmic microtubules length by Bik1, Kip2, Kip3, and not on a capture/shrinkage mechanism.
Grava S, Philippsen P. Grava S, et al. Mol Biol Cell. 2010 Nov 1;21(21):3680-92. doi: 10.1091/mbc.E10-06-0527. Epub 2010 Sep 15. Mol Biol Cell. 2010. PMID: 20844079 Free PMC article. - Cytoplasmic dynein and early endosome transport.
Xiang X, Qiu R, Yao X, Arst HN Jr, Peñalva MA, Zhang J. Xiang X, et al. Cell Mol Life Sci. 2015 Sep;72(17):3267-80. doi: 10.1007/s00018-015-1926-y. Epub 2015 May 23. Cell Mol Life Sci. 2015. PMID: 26001903 Free PMC article. Review. - Point mutations in the stem region and the fourth AAA domain of cytoplasmic dynein heavy chain partially suppress the phenotype of NUDF/LIS1 loss in Aspergillus nidulans.
Zhuang L, Zhang J, Xiang X. Zhuang L, et al. Genetics. 2007 Mar;175(3):1185-96. doi: 10.1534/genetics.106.069013. Epub 2007 Jan 21. Genetics. 2007. PMID: 17237507 Free PMC article. - Aspergillus nidulans Dis1/XMAP215 protein AlpA localizes to spindle pole bodies and microtubule plus ends and contributes to growth directionality.
Enke C, Zekert N, Veith D, Schaaf C, Konzack S, Fischer R. Enke C, et al. Eukaryot Cell. 2007 Mar;6(3):555-62. doi: 10.1128/EC.00266-06. Epub 2007 Jan 19. Eukaryot Cell. 2007. PMID: 17237365 Free PMC article. - The splicing-factor Prp40 affects dynein-dynactin function in Aspergillus nidulans.
Qiu R, Zhang J, Xiang X. Qiu R, et al. Mol Biol Cell. 2020 Jun 1;31(12):1289-1301. doi: 10.1091/mbc.E20-03-0166. Epub 2020 Apr 8. Mol Biol Cell. 2020. PMID: 32267207 Free PMC article.
References
- Arnal, I., Heichette, C., Diamantopoulos, G. S., and Chretien, D. (2004). CLIP-170/tubulin-curved oligomers coassemble at microtubule ends and promote rescues. Curr. Biol. 14, 2086-2095. - PubMed
- Akhmanova, A., and Hoogenraad, C. C. (2005). Microtubule plus-end-tracking proteins: mechanisms and functions. Curr. Opin. Cell Biol. 17, 47-54. - PubMed
- Bloom, K. (2005). Chromosome segregation: seeing is believing. Curr. Biol. 15, R500-R503. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous