Impaired neuronal positioning and dendritogenesis in the neocortex after cell-autonomous Dab1 suppression - PubMed (original) (raw)
Impaired neuronal positioning and dendritogenesis in the neocortex after cell-autonomous Dab1 suppression
Eric C Olson et al. J Neurosci. 2006.
Abstract
Reelin and Disabled 1 (Dab1) are essential for positioning migrating neurons in the developing neocortex. Cell-autonomous RNA interference-mediated suppression of Dab1 in migrating neurons destined for layer 2/3 shifted the median position of these cells to deeper positions within the cortex. At the time of migration arrest [embryonic day 20 (E20) to E21], Dab1-suppressed cells were underrepresented in the upper approximately 40 microm of the cortex compared with controls, suggesting that Dab1 is essential for somal translocation through the cell-dense cortical plate. Closer examination of the morphology of Dab1-suppressed neurons at E20 revealed simplified leading processes that are less likely to contact the marginal zone (MZ), in which high levels of Reelin are expressed. Examination of Dab1-suppressed cells 3 d later (postnatal day 2) revealed simplified dendrites that are also less likely to contact the MZ. These data reveal a cell-autonomous role of Dab1 in dendritogenesis in the neocortex and suggest that remodeling of the leading process of a migrating neuron into a nascent dendrite by Reelin/Dab1 signaling plays an important role in cell positioning.
Figures
Figure 1.
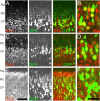
Expression of layer-appropriate markers in electroporated cells. P0 coronal sections of cerebral cortex 5 d after E16 pCAG-GFP electroporations show appropriate expression of layer-specific marker. GFP+ cells in the P0 cortex immunolocalize with the POU domain transcription factor Brn1, a marker of layer 2–5 cells (A, B), and the Cut-related transcription factor Cux1, a marker of layer 2–4 cells (C, D) in the mouse cortex. In addition, GFP+ cells are positive for DCX, a marker of immature postmitotic neurons (E,F). B,D, and F are single confocal sections that have been digitally zoomed three times, whereas**A, C, andE** are flattened _z_-series through 10 μm of tissue. Scale bars: E, 50 μm;F, 20 μm.
Figure 2.
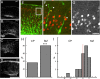
In vivo morphological changes in migrating neurons in contact with the marginal zone. Time course of cell migration after E16 electroporation.A, At E17, 1 d after electroporation, GFP+ cells in sections of the neocortex are found primarily in the ventricular zone (VZ). By E19 (B) and E20 (C), GFP+ cells have entered the cortex and are approaching the MZ. D, By P0, typically 5 d after electroporation, GFP+ cells are found in a tight layer underneath the MZ. E, F, Examples of the first group of GFP+ cells contacting the MZ from an E19 section of E16 pCAG-GFP-electroporated (green) and E16 bromodeoxyuridine (BrdU)-labeled (red) cortex. E, The white box identifies the migrating cells at the leading edge of the GFP+ cells that are examined at higher magnification in F. The cell on the left has a clear branch (yellow arrow) approximately two cell diameters above the CP, but the cell on the right is not branched. G, Arrows identify the BrdU+ nuclei from the cells shown in F.H, A higher percentage of cells show leading process branching if the process was in the MZ as opposed to the cortex (CP) (***p < 0.001, χ2 test; n = 30 leading processes in MZ; n = 25 leading processes in CP).I, Histogram of the location of the principal branch point with respect to the CP–MZ boundary (red line, _x_= 0). The _x_-axis is in nuclear diameters (7 μm). A majority of branch points in MZ-contacting cells were found in the first two cell diameters above the CP–MZ boundary. The leading cell population (n = 55 cells) was derived from _z_-series from four E16 electroporated brains fixed at E19 (n = 1) or E20 (n = 3). Scale bars: D, 500 μm;G, 25 μm.
Figure 3.
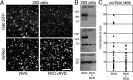
Suppression of Dab1 protein expression by RNAi directed against Dab1 mRNA.A, HEK293 cells cotransfected with pSuper.retro.neo plasmids directed against Dab1 (RVC+RVD) and with a plasmid encoding Dab1::GFP show reduced the amount of Dab1::GFP expression compared with control transfection (RVN with Dab1::GFP). A plasmid encoding the red fluorescent protein DsRed (pDsRed) was included in all transfections as a transfection efficiency control. Three independent transfections of HEK293 cell cultures were performed to test Dab1::GFP suppression by RVN, RVC, and RVD. Green to red fluorescence ratios within regions of interest was 1.05 ± 0.25 for RVN-treated cultures (n = 3) compared with 0.20 ± 0.23 for RVC+RVD-treated cultures (n = 3) (means ± SD; unpaired _t_test, p = 0.012; df = 4). B, Western blot analysis of whole-cell lysates from HEK293 cell transfections confirms Dab1::GFP suppression after probing with anti-Dab1 antisera (B3) or with an anti-GFP antibody. Reprobing the immunoblot with an anti-α-tubulin antibody confirms approximately equal loading of whole-cell lysates into each lane.C, Suppression of endogenous Dab1 in primary cortical cultures after electroporation of RNAi constructs with pCAG-GFP. Quantification of Dab1 immunostaining in the GFP+ populations in control (RVN) and (RVC+RVD) electroporated cell cultures. Mean pixel intensities of the cell soma are expressed in arbitrary units (au) after background correction (subtraction of the mean pixel intensity from a cell-free region of the same culture). A difference in mean pixel intensity is observed between RVN- and RVC+RVD-treated neuronal cultures (p = 0.001, unpaired t test; df = 58; n = 30 GFP+ cells in each condition). Neurons derived from three electroporated and pooled brains were quantified in each condition in C. Scale bar, 230 μm.
Figure 4.
Dispersed cellular layering after treatment with RNAi directed against Dab1. Sections stained with the nucleic acid dye propidium iodide (red) from embryos electroporated on E16 and examined on E20 or E21 in the cingulate (A) or lateral neocortex (B). RVN control electroporated brains show precise layering by E20 and E21. B, Embryos electroporated with RNAi plasmids (RVC+RVD) directed against Dab1 show more cellular dispersion and fewer cells at the CP–MZ boundary. Scale bars: low-magnification images, 250 μm; high-magnification images, 50 μm. C,D, Binned histogram of the distribution of GFP+ cell bodies in the upper 320 μm of the cingulate cortex (C) and lateral neocortex (D). Control RVN electroporations (black bars) show more cells in the upper bins (bins 1–2) than RVC+RVD (white bars). Coelectroporation of a cDNA encoding full-length Dab1 (cross-hatched bars) partially rescues RVC+RVD malpositioning. A total of 48 electroporated brains were examined, six to nine brains for each condition at each time point in the lateral neocortex, two to four brains for each condition at each time point in the cingulate cortex, and four brains for the Dab1 rescue in the lateral neocortex at E21. The soma positions for a total of 2792 cells or an average of 58 cells per brain were measured. CTX, Cortex.
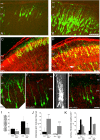
Figure 5.
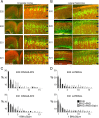
Alterations of leading process and apical dendrite morphology after treatment with RNAi directed against Dab1. Sections stained with the nucleic acid dye propidium iodide (red) from embryos electroporated at E16 and examined at E20 and P2.A, Control (RVN) neurons electroporated at E16 show branched neurites at E20. B, RVC+RVD-treated neurons show less branching in the apical process contacting the MZ.C, D, Low-magnification images of layer 2/3 cortical neurons on P2, 7 d after in utero electroporation on E16. Control RVN electroporated neurons show precise lamination and exuberant dendritic growth in the MZ on P2 (C), whereas Dab1-suppressed cells (RVC+RVD) show disrupted lamination with occasional ectopic deep cells (arrow) and sparse dendrites in the MZ (D).C–F, Flattened z-series of P2 layer 2/3 neurons revealing extensive dendrites in the RVN-treated cells (E) and stunted dendrites that either do not penetrate the MZ (cells 2 and 3) (F–H) or stunted dendrites that do not show extensive secondary and tertiary branching in the MZ (H). G, A 90° rotation of D reveals that cells 1–3 are entirely contained in the optical section and that the stunted dendrites are not attributable to sectioning artifact. Scale bars: B,D, 50 μm; H, 20 μm. I, Quantification of GFP+pixels in the MZ at E20 and P2, after normalization (see Materials and Methods). Control (RVN) embryos showed approximately twofold more neurites in the MZ compared with RVC+RVD-treated embryos at both E20 (p = 0.014, unpaired_t_ test; df = 6) and P2 (p = 0.025, unpaired_t_ test; df = 7). Single sections from nine RVC+RVD embryos (n = 4 at E20; n = 5 at P2) and eight RVN embryos (n = 4 at E20; n = 4 at P2) were analyzed.J, Quantification of noncontacting apical processes after RVC+RVD treatment shows RNAi-mediated difference at both E20 (p = 0.012, unpaired t test; df = 4) and P2 (p = 0.001, unpaired t test; df = 6).K, Quantification of apical process branching complexity from unbranched (0th order) to second-order and above (2+) branching in RVN- and RVC+RVD-treated cells at E20 and P2. RVC+RVD-treated cells showed less branching than RVN-treated cells at both time points. At E20, morphological analysis was performed on 24 cells in three RVN-treated brains and 26 cells in three RVC+RVD-treated brains. At P2, morphological analysis was performed on 47 cells in five RVN-treated brains and 44 cells in three RVC+RVD-treated brains. Values in**I** and J are means ± SD. CTX, Cortex.
Similar articles
- Ectopic Reelin induces neuronal aggregation with a normal birthdate-dependent "inside-out" alignment in the developing neocortex.
Kubo K, Honda T, Tomita K, Sekine K, Ishii K, Uto A, Kobayashi K, Tabata H, Nakajima K. Kubo K, et al. J Neurosci. 2010 Aug 18;30(33):10953-66. doi: 10.1523/JNEUROSCI.0486-10.2010. J Neurosci. 2010. PMID: 20720102 Free PMC article. - Alternative Splicing of Disabled-1 Controls Multipolar-to-Bipolar Transition of Migrating Neurons in the Neocortex.
Zhang B, Wang W, Zhang Z, Hu Y, Meng F, Wang F, Lou H, Zhu L, Godbout R, Duan S, Gao Z. Zhang B, et al. Cereb Cortex. 2018 Oct 1;28(10):3457-3467. doi: 10.1093/cercor/bhx212. Cereb Cortex. 2018. PMID: 28968791 - Neurons tend to stop migration and differentiate along the cortical internal plexiform zones in the Reelin signal-deficient mice.
Tabata H, Nakajima K. Tabata H, et al. J Neurosci Res. 2002 Sep 15;69(6):723-30. doi: 10.1002/jnr.10345. J Neurosci Res. 2002. PMID: 12205665 - How does Reelin control neuronal migration and layer formation in the developing mammalian neocortex?
Sekine K, Kubo K, Nakajima K. Sekine K, et al. Neurosci Res. 2014 Sep;86:50-8. doi: 10.1016/j.neures.2014.06.004. Epub 2014 Jun 23. Neurosci Res. 2014. PMID: 24969097 Review. - [Corticohistogenesis and Reelin signal cascade].
Ogawa M. Ogawa M. Nihon Shinkei Seishin Yakurigaku Zasshi. 2000 Oct;20(4):169-74. Nihon Shinkei Seishin Yakurigaku Zasshi. 2000. PMID: 11215402 Review. Japanese.
Cited by
- Impaired structural and functional development of cerebellum following gestational exposure of deltamethrin in rats: role of reelin.
Kumar K, Patro N, Patro I. Kumar K, et al. Cell Mol Neurobiol. 2013 Jul;33(5):731-46. doi: 10.1007/s10571-013-9942-7. Epub 2013 May 17. Cell Mol Neurobiol. 2013. PMID: 23681596 Free PMC article. - Combined transcriptome analysis of fetal human and mouse cerebral cortex exposed to alcohol.
Hashimoto-Torii K, Kawasawa YI, Kuhn A, Rakic P. Hashimoto-Torii K, et al. Proc Natl Acad Sci U S A. 2011 Mar 8;108(10):4212-7. doi: 10.1073/pnas.1100903108. Epub 2011 Feb 22. Proc Natl Acad Sci U S A. 2011. PMID: 21368140 Free PMC article. - Ectopic Reelin induces neuronal aggregation with a normal birthdate-dependent "inside-out" alignment in the developing neocortex.
Kubo K, Honda T, Tomita K, Sekine K, Ishii K, Uto A, Kobayashi K, Tabata H, Nakajima K. Kubo K, et al. J Neurosci. 2010 Aug 18;30(33):10953-66. doi: 10.1523/JNEUROSCI.0486-10.2010. J Neurosci. 2010. PMID: 20720102 Free PMC article. - A critical function for beta-amyloid precursor protein in neuronal migration revealed by in utero RNA interference.
Young-Pearse TL, Bai J, Chang R, Zheng JB, LoTurco JJ, Selkoe DJ. Young-Pearse TL, et al. J Neurosci. 2007 Dec 26;27(52):14459-69. doi: 10.1523/JNEUROSCI.4701-07.2007. J Neurosci. 2007. PMID: 18160654 Free PMC article. - Reelin stabilizes the actin cytoskeleton of neuronal processes by inducing n-cofilin phosphorylation at serine3.
Chai X, Förster E, Zhao S, Bock HH, Frotscher M. Chai X, et al. J Neurosci. 2009 Jan 7;29(1):288-99. doi: 10.1523/JNEUROSCI.2934-08.2009. J Neurosci. 2009. PMID: 19129405 Free PMC article.
References
- Caviness VS Jr, Sidman RL (1973). Time of origin or corresponding cell classes in the cerebral cortex of normal and reeler mutant mice: an autoradiographic analysis. J Comp Neurol 148:141–151. - PubMed
- D’Apuzzo M, Mandolesi G, Reis G, Schuman EM (2001). Abundant GFP expression and LTP in hippocampal acute slices by in vivo injection of sindbis virus. J Neurophysiol 86:1037–1042. - PubMed
- D’Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, Curran T (1995). A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature 374:719–723. - PubMed
- D’Arcangelo G, Homayouni R, Keshvara L, Rice DS, Sheldon M, Curran T (1999). Reelin is a ligand for lipoprotein receptors. Neuron 24:471–479. - PubMed
- Derer P, Derer M, Goffinet A (2001). Axonal secretion of Reelin by Cajal-Retzius cells: evidence from comparison of normal and Reln(Orl) mutant mice. J Comp Neurol 440:136–143. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources