DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation - PubMed (original) (raw)
DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation
Mary Gehring et al. Cell. 2006.
Abstract
MEDEA (MEA) is an Arabidopsis Polycomb group gene that is imprinted in the endosperm. The maternal allele is expressed and the paternal allele is silent. MEA is controlled by DEMETER (DME), a DNA glycosylase required to activate MEA expression, and METHYLTRANSFERASE I (MET1), which maintains CG methylation at the MEA locus. Here we show that DME is responsible for endosperm maternal-allele-specific hypomethylation at the MEA gene. DME can excise 5-methylcytosine in vitro and when expressed in E. coli. Abasic sites opposite 5-methylcytosine inhibit DME activity and might prevent DME from generating double-stranded DNA breaks. Unexpectedly, paternal-allele silencing is not controlled by DNA methylation. Rather, Polycomb group proteins that are expressed from the maternal genome, including MEA, control paternal MEA silencing. Thus, DME establishes MEA imprinting by removing 5-methylcytosine to activate the maternal allele. MEA imprinting is subsequently maintained in the endosperm by maternal MEA silencing the paternal allele.
Figures
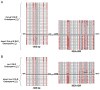
Figure 1. MEA Methylation in Dissected Seeds
(A) MEA is methylated in four regions. Numbers are relative to the translation start site. (B) CG methylation of maternal and paternal embryo and endosperm alleles from a Col-gl female crossed to a RLD male. The number of clones sequenced is given at the base of each column. Black lines, sequences assayed by bisulfite sequencing; blue bar, helitron transposon element; red arrowheads, 182 bp direct repeats; lollipops, sites of DNA methylation (red, CG; blue, CNG; gray, CNN).
Figure 2. Hypermethylation of Maternal MEA in dme Mutant Endosperm
Maternal-allele methylation in the −500 bp and MEA-ISR regions in endosperm from crosses between dme-2 heterozygous females and RLD males compared to maternal endosperm allele methylation from crosses between wild-type females and RLD males. (A) dme-2 heterozygous Col-gl crossed to RLD. (B) dme-2 heterozygous L_er_ crossed to RLD. Mutant endosperm was collected at 9 DAP from seeds with the dme endosperm overproliferation phenotype. Numbers are from the translation start site. To determine the pattern of DNA methylation, DNA was treated with bisulfite, PCR amplified, cloned, and sequenced. Circles connected by lines represent the results from determining the DNA sequence of one clone. Filled circle, methylated cytosine; open circle, unmethylated cytosine; red circle, CG site; blue circle, CNG site; gray circle, CNN site.
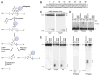
Figure 3. DME In Vitro Activity
(A) Schematic mechanism of bifunctional DNA glycosylases. (B) DNA substrate sequence. Base pair positions relative to the 5′ end of the top DNA strand are shown. Double-stranded DNA oligonucleotide substrates in (C)–(E) were labeled at the 5′ end of the top strand. DNAs in (C) had 5-methylcytosine at position 18 in the top strand. The top strand for (D) and (E) has: CpG, C at position 18; meCpG, 5-methylcytosine at position 18; T/G, T at position 18; meCpNpG, 5-methylcytosine at position 17; meCpNpN, 5-methylcytosine at position 15. All reactions were for 1 hr. (C) Reaction products of DME. Products were treated with either water or NaOH as indicated, denatured, and analyzed on 15% polyacrylamide gels with 7.5 M urea. (D) Covalent crosslinking of DME to DNA. Reaction products were treated with NaBH4, denatured, and analyzed on a 10% SDS-polyacrylamide gel. (E) Substrate specificity of DME. Reaction products were denatured and analyzed on 15% polyacrylamide gels with 7.5 M urea. Both β and δ elimination products are observed because reactions were not treated with NaOH before gel electrophoresis. S, uncleaved substrate; β, predicted β elimination product; δ, predicted δ elimination product; 35 nt, 35 nucleotide size marker; 17 nt, 17 nucleotide size marker.
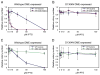
Figure 4. DME Functions as a 5-methylcytosine DNA Glycosylase in E. coli
Relative colony number; number of colonies on plate divided by the number of colonies obtained when plate has no IPTG inducer. The relative colony number at each concentration of IPTG was determined three times. Error bars represent the standard deviation from the mean. (A and B) wt bacteria, AB1157; AP endo mutant RPC501 (Cunningham et al., 1986) isogenic to AB1157, with mutations in two AP endonuclease genes (xth, nfo). (C and D) wt bacteria, GM30; DNA met mutant GM31 (Palmer and Marinus, 1994) isogenic to GM30, with a mutation in the dcm DNA methyltransferase.
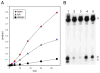
Figure 5. Inhibition of DME Activity by Abasic Sites
(A) Rate of DME activity. Labeled (5′ end of the bottom strand) double-stranded oligonucleotides (Figure 3B) were used with the following sequences: hemi, 5-methylcytosine at position 19 (bottom strand); full, 5-methylcytosine at positions 19 (bottom strand) and 18 (top strand); abasic, 5-methylcytosine at position 19 (bottom strand) and an abasic site at 18 (top strand). Reactions were performed, terminated by addition of NaOH, boiled, and subjected to electrophoresis. Gels were exposed to a phosphorimager screen to determine the amount of product. (B) Effect of abasic-site position on DME activity. Double-stranded oligonucleotides (Figure 3B) were labeled at the 5′ end of the bottom strand and had 5-methylcytosine at position 19 of the bottom strand (lane 1). In addition, abasic sites were in the top strand at position 18 (lane 2), position 17 (lane 3), position 15 (lane 4), and position 12 (lane 5).
Figure 6. Regulation of MEA Paternal-Allele Silencing
(A) Paternal MEA silencing is not affected by a hypomethylated paternal genome. Expression of MEA in the embryo and endosperm/seed coat of crosses between a RLD female and Col-gl male and a RLD female and a met1-6 homozygous Col-gl male. Seeds were dissected 7 DAP. (B) MEA expression in mutant endosperm of crosses between mea-3 homozygous L_er_, fie-1 heterozygous L_er_, and dme-2 heterozygous Col-gl females and RLD males, dissected 9 DAP. (C) MEA expression in endosperm of crosses between L_er_ and mea-3 homozygous L_er_ females and Cvi males, dissected 7 and 8 DAP, respectively, at the torpedo stage of embryogenesis. VPE is a control for biallelic expression. (D) Genomic structure of ArabidopsisMEA_andregions examinedbyChIP.E1 throughE4;exons 1 through4.Regions amplified are shown bybars labeled 1 and 2. (E) ChIP with anti-dimethylH3K27 comparing amplification of_MEA in wt L_er_ X RLD andmutant L_ermea_ X RLD siliques 7 DAP. LNA primers were used to amplify regions 1 and 2 and not the actin control DNA.
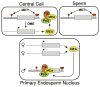
Figure 7. Model for Regulation of MEA Imprinting
MEA methylation is maintained by MET1. In the central cell, DME removes methylation at the −500 bp region and MEA-ISR. MEA protein is produced and forms PcG complexes. After fertilization, MEA-FIE PcG complexes target the paternal allele to maintain its silent state. Maternal MEA continues to be expressed in the endosperm. Gray box, MEA gene; red circles, DNA methylation; helical line, nontranscribed compacted chromatin; straight line, transcribed open chromatin.
Comment in
- MEDEA takes control of its own imprinting.
Arnaud P, Feil R. Arnaud P, et al. Cell. 2006 Feb 10;124(3):468-70. doi: 10.1016/j.cell.2006.01.020. Cell. 2006. PMID: 16469694
Similar articles
- Imprinting of the MEA Polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase.
Xiao W, Gehring M, Choi Y, Margossian L, Pu H, Harada JJ, Goldberg RB, Pennell RI, Fischer RL. Xiao W, et al. Dev Cell. 2003 Dec;5(6):891-901. doi: 10.1016/s1534-5807(03)00361-7. Dev Cell. 2003. PMID: 14667411 - Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte.
Schoft VK, Chumak N, Choi Y, Hannon M, Garcia-Aguilar M, Machlicova A, Slusarz L, Mosiolek M, Park JS, Park GT, Fischer RL, Tamaru H. Schoft VK, et al. Proc Natl Acad Sci U S A. 2011 May 10;108(19):8042-7. doi: 10.1073/pnas.1105117108. Epub 2011 Apr 25. Proc Natl Acad Sci U S A. 2011. PMID: 21518889 Free PMC article. - Histone H1 affects gene imprinting and DNA methylation in Arabidopsis.
Rea M, Zheng W, Chen M, Braud C, Bhangu D, Rognan TN, Xiao W. Rea M, et al. Plant J. 2012 Sep;71(5):776-86. doi: 10.1111/j.1365-313X.2012.05028.x. Epub 2012 Jun 14. Plant J. 2012. PMID: 22519754 Free PMC article. - Imprinting in plants and its underlying mechanisms.
Zhang H, Chaudhury A, Wu X. Zhang H, et al. J Genet Genomics. 2013 May 20;40(5):239-47. doi: 10.1016/j.jgg.2013.04.003. Epub 2013 Apr 20. J Genet Genomics. 2013. PMID: 23706299 Review. - Genome demethylation and imprinting in the endosperm.
Bauer MJ, Fischer RL. Bauer MJ, et al. Curr Opin Plant Biol. 2011 Apr;14(2):162-7. doi: 10.1016/j.pbi.2011.02.006. Epub 2011 Mar 23. Curr Opin Plant Biol. 2011. PMID: 21435940 Free PMC article. Review.
Cited by
- Recent advances in the structural mechanisms of DNA glycosylases.
Brooks SC, Adhikary S, Rubinson EH, Eichman BF. Brooks SC, et al. Biochim Biophys Acta. 2013 Jan;1834(1):247-71. doi: 10.1016/j.bbapap.2012.10.005. Epub 2012 Oct 14. Biochim Biophys Acta. 2013. PMID: 23076011 Free PMC article. Review. - Mutation of the imprinted gene OsEMF2a induces autonomous endosperm development and delayed cellularization in rice.
Tonosaki K, Ono A, Kunisada M, Nishino M, Nagata H, Sakamoto S, Kijima ST, Furuumi H, Nonomura KI, Sato Y, Ohme-Takagi M, Endo M, Comai L, Hatakeyama K, Kawakatsu T, Kinoshita T. Tonosaki K, et al. Plant Cell. 2021 Mar 22;33(1):85-103. doi: 10.1093/plcell/koaa006. Plant Cell. 2021. PMID: 33751094 Free PMC article. - Methyl-CpG-binding domain protein MBD7 is required for active DNA demethylation in Arabidopsis.
Wang C, Dong X, Jin D, Zhao Y, Xie S, Li X, He X, Lang Z, Lai J, Zhu JK, Gong Z. Wang C, et al. Plant Physiol. 2015 Mar;167(3):905-14. doi: 10.1104/pp.114.252106. Epub 2015 Jan 15. Plant Physiol. 2015. PMID: 25593350 Free PMC article. - The cytosolic Fe-S cluster assembly component MET18 is required for the full enzymatic activity of ROS1 in active DNA demethylation.
Wang X, Li Q, Yuan W, Cao Z, Qi B, Kumar S, Li Y, Qian W. Wang X, et al. Sci Rep. 2016 May 19;6:26443. doi: 10.1038/srep26443. Sci Rep. 2016. PMID: 27193999 Free PMC article. - Parent-of-origin effects on gene expression and DNA methylation in the maize endosperm.
Waters AJ, Makarevitch I, Eichten SR, Swanson-Wagner RA, Yeh CT, Xu W, Schnable PS, Vaughn MW, Gehring M, Springer NM. Waters AJ, et al. Plant Cell. 2011 Dec;23(12):4221-33. doi: 10.1105/tpc.111.092668. Epub 2011 Dec 23. Plant Cell. 2011. PMID: 22198147 Free PMC article.
References
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427:164–167. - PubMed
- Bender J. DNA methylation and epigenetics. Annu Rev Plant Biol. 2004;55:41–68. - PubMed
- Bhagwat M, Gerlt JA. 3′- and 5′-strand cleavage reactions catalyzed by the Fpg protein from Escherichia coli occur via successive β- and δ-elimination mechanisms, respectively. Biochemistry. 1996;35:659–665. - PubMed
- Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–164. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases