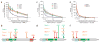Feedback repression is required for mammalian circadian clock function - PubMed (original) (raw)
Feedback repression is required for mammalian circadian clock function
Trey K Sato et al. Nat Genet. 2006 Mar.
Abstract
Direct evidence for the requirement of transcriptional feedback repression in circadian clock function has been elusive. Here, we developed a molecular genetic screen in mammalian cells to identify mutants of the circadian transcriptional activators CLOCK and BMAL1, which were uncoupled from CRYPTOCHROME (CRY)-mediated transcriptional repression. Notably, mutations in the PER-ARNT-SIM domain of CLOCK and the C terminus of BMAL1 resulted in synergistic insensitivity through reduced physical interactions with CRY. Coexpression of these mutant proteins in cultured fibroblasts caused arrhythmic phenotypes in population and single-cell assays. These data demonstrate that CRY-mediated repression of the CLOCK/BMAL1 complex activity is required for maintenance of circadian rhythmicity and provide formal proof that transcriptional feedback is required for mammalian clock function.
Figures
Figure 1
Mutations in CLOCK and BMAL1 confer insensitivity to CRY-mediated transcriptional repression without affecting CLOCK/BMAL1 transcriptional activity. (a,c,e) Results of cell-based transcriptional _PER1_-luciferase reporter assays with mutant CLOCK and BMAL1 clones in HEK293T cells. Plasmids expressing Flag-tagged wild-type or mutant CLOCK (a) or BMAL1 (c,e) cDNAs were transiently cotransfected with wild-type (WT) BMAL1 or CLOCK, respectively, _PER1_-luciferase reporter and 0–5 ng of CRY1 plasmid. Activity is expressed as the percentage of normalized _PER1_-luciferase activity in cells transfected with wild-type CLOCK/BMAL1 alone. Data are mean ± s.e.m. from independent experimental triplicates. (b,d,f) Domain locations of causative mutations. Schematic locations of amino acid changes within the CLOCK PAS-B domain (b) and BMAL1 C terminus (d) that confer CRY1 desensitization are indicated. Locations of protein domains are indicated for CLOCK (bHLH, blue, amino acids 35–85; PAS domain, green, amino acids 113–377; PAS-A repeat, orange, amino acids 128–170; PAS-B repeat, yellow, amino acids 283–329) and BMAL1 (bHLH, blue, amino acids 73–126; PAS domain, green, amino acids 148–439; PAS-A repeat, orange, amino acids 163–206; PAS-B repeat, yellow, amino acids 344–391). (f) Schematic location of amino acid changes within the N terminus of BMAL1 that confer CLOCK/BMAL1 hyperactivity: Bmal1-5, S10L and V160I; Bmal1-6, S9F.
Figure 2
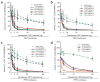
Coexpression of CLOCK and BMAL1 desensitized mutants confers synergistic insensitivity to CRY1 in HEK293T cells. (a–c) Various combinations of single and double CRY-desensitized CLOCK and BMAL1 mutant cDNAs were cotransfected with the PER1 reporter and 0–50 ng of CRY1 plasmid. PER1 reporter activity alone (blue triangle) or with wild-type CLOCK/BMAL1 (orange triangle) are also displayed. Activity is expressed as the percentage of normalized _PER1_-luciferase activity in cells transfected with wild-type CLOCK/BMAL1 alone. Solid lines: CLOCK and BMAL1 single mutants. Dashed lines: double mutants. (d) Luciferase activities were analyzed from combinations of single and double Bmal1-3 and Clock-3 mutant cDNAs that were cotransfected with the PER1 reporter and 0–25 ng of CRY2 plasmid. PER1 reporter activity alone (blue triangle) or with wild-type CLOCK/BMAL1 (green triangle) are also shown. Activity is expressed as the percentage of normalized _PER1_-luciferase activity in transfection with wild-type CLOCK/BMAL1 and no CRY1 plasmid. Data are mean ± s.e.m. determined from independent experimental triplicates.
Figure 3
Mutations in the CLOCK PAS domain and BMAL1 C terminus abrogate interactions between the CLOCK/BMAL1 complex and transcriptional repressors CRY1 and PER2. (a–c) Native coimmunoprecipitations (co-IPs) were performed on cell extracts from HEK293T cells transiently transfected with plasmids expressing epitope-tagged circadian components. (a) Flag-tagged wild-type or mutant BMAL1 protein was coexpressed with or without CRY1 and various untagged CLOCK alleles and precipitated with anti-Flag. Copurified proteins were visualized by protein blotting with antibodies to Flag or CLOCK. (b) Co-IPs were performed with anti-MYC on extracts from cells coexpressing MYC-tagged CRY1, combinations of various Flag-tagged CLOCK and BMAL1 CRY-desensitized alleles and Flag-tagged PER2. Anti-Flag and CRY1 antibodies were used in protein blotting. (c) Flag-PER2 was coprecipitated (using anti-Flag) from cells coexpressing additional combinations of untagged CLOCK and BMAL1 alleles. Protein blotting with anti-Flag, anti-CLOCK and anti-BMAL1 was used to identify copurified proteins.
Figure 4
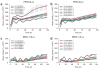
Coexpression of CLOCK/BMAL1 mutant heterodimers that are insensitive to CRY repression ablates circadian E-box and RORE activities in NIH3T3 cells. Plasmids expressing Flag-tagged CLOCK and BMAL1 alleles were transiently cotransfected with the _PER2_-destabilized luciferase (dLuc) reporter plasmid into NIH3T3 cells. (a,b) PER2 promoter activities in NIH3T3 cells transfected with single (a) or double (b) CRY1-insensitive CLOCK and BMAL1 mutants were monitored over 5 d. (c,d) BMAL1 promoter activities in NIH3T3 cells transfected with single (c) or double (d) CRY-insensitive mutants of CLOCK and BMAL1 were monitored over 6 d. All reporter activities were normalized such that the median wild-type luciferase activity over the time-course was 100%.
Figure 5
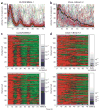
Coexpression of CLOCK/BMAL1 mutant heterodimers impairs circadian rhythmicity in individual cells. (a–d) _PER2_-luciferase reporter activities from individual NIH3T3 cells (n = 133) transfected with Flag-tagged wild-type CLOCK/BMAL1 (a,c) or double-mutant Clock-1/Bmal1-3 (b,d) were monitored over 3 d. Reporter activities from each wild-type (a) or double-mutant (b) cell were normalized such that the maximum bioluminescence value was set to 100% for each panel. The mean reporter activity for all analyzed single cells at each time point is indicated by a black line. Normalized bioluminescence activities from each wild-type (c) or double-mutant (d) cell were detrended and ranked according to corresponding significance by autocorrelation (upper panels) or COSOPT (lower panels). Autocorrelation and MMC-β values for each cell are depicted to the right of each heat map from 0 (bottom, dark blue) to 1 (top, white). Red and green denote high and low normalized reporter activities, respectively. Results shown are representative of duplicate experiments.
Similar articles
- Dual role of the CLOCK/BMAL1 circadian complex in transcriptional regulation.
Kondratov RV, Shamanna RK, Kondratova AA, Gorbacheva VY, Antoch MP. Kondratov RV, et al. FASEB J. 2006 Mar;20(3):530-2. doi: 10.1096/fj.05-5321fje. Epub 2006 Jan 25. FASEB J. 2006. PMID: 16507766 - The BMAL1 C terminus regulates the circadian transcription feedback loop.
Kiyohara YB, Tagao S, Tamanini F, Morita A, Sugisawa Y, Yasuda M, Yamanaka I, Ueda HR, van der Horst GT, Kondo T, Yagita K. Kiyohara YB, et al. Proc Natl Acad Sci U S A. 2006 Jun 27;103(26):10074-9. doi: 10.1073/pnas.0601416103. Epub 2006 Jun 15. Proc Natl Acad Sci U S A. 2006. PMID: 16777965 Free PMC article. - BMAL1-dependent circadian oscillation of nuclear CLOCK: posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system.
Kondratov RV, Chernov MV, Kondratova AA, Gorbacheva VY, Gudkov AV, Antoch MP. Kondratov RV, et al. Genes Dev. 2003 Aug 1;17(15):1921-32. doi: 10.1101/gad.1099503. Genes Dev. 2003. PMID: 12897057 Free PMC article. - Genetics and neurobiology of circadian clocks in mammals.
Siepka SM, Yoo SH, Park J, Lee C, Takahashi JS. Siepka SM, et al. Cold Spring Harb Symp Quant Biol. 2007;72:251-259. doi: 10.1101/sqb.2007.72.052. Cold Spring Harb Symp Quant Biol. 2007. PMID: 18419282 Free PMC article. Review. - [Synchronization and genetic redundancy in circadian clocks].
Dardente H. Dardente H. Med Sci (Paris). 2008 Mar;24(3):270-6. doi: 10.1051/medsci/2008243270. Med Sci (Paris). 2008. PMID: 18334175 Review. French.
Cited by
- The mammalian circadian system is resistant to dioxin.
Pendergast JS, Yamazaki S. Pendergast JS, et al. J Biol Rhythms. 2012 Apr;27(2):156-63. doi: 10.1177/0748730411434405. J Biol Rhythms. 2012. PMID: 22476776 Free PMC article. - Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex.
Huang N, Chelliah Y, Shan Y, Taylor CA, Yoo SH, Partch C, Green CB, Zhang H, Takahashi JS. Huang N, et al. Science. 2012 Jul 13;337(6091):189-94. doi: 10.1126/science.1222804. Epub 2012 May 31. Science. 2012. PMID: 22653727 Free PMC article. - The human CRY1 tail controls circadian timing by regulating its association with CLOCK:BMAL1.
Parico GCG, Perez I, Fribourgh JL, Hernandez BN, Lee HW, Partch CL. Parico GCG, et al. Proc Natl Acad Sci U S A. 2020 Nov 10;117(45):27971-27979. doi: 10.1073/pnas.1920653117. Epub 2020 Oct 26. Proc Natl Acad Sci U S A. 2020. PMID: 33106415 Free PMC article. - BMAL1 regulates _Propionibacterium acnes_-induced skin inflammation via REV-ERBα in mice.
Li F, Lin L, He Y, Sun G, Dong D, Wu B. Li F, et al. Int J Biol Sci. 2022 Mar 21;18(6):2597-2608. doi: 10.7150/ijbs.71719. eCollection 2022. Int J Biol Sci. 2022. PMID: 35414779 Free PMC article. - High-throughput and single-cell imaging of NF-kappaB oscillations using monoclonal cell lines.
Bartfeld S, Hess S, Bauer B, Machuy N, Ogilvie LA, Schuchhardt J, Meyer TF. Bartfeld S, et al. BMC Cell Biol. 2010 Mar 16;11:21. doi: 10.1186/1471-2121-11-21. BMC Cell Biol. 2010. PMID: 20233427 Free PMC article.
References
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. - PubMed
- Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. - PubMed
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. - PubMed
- Nakajima M, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. - PubMed
- Tomita J, Nakajima M, Kondo T, Iwasaki H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307:251–254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases