Crystal structure of human indoleamine 2,3-dioxygenase: catalytic mechanism of O2 incorporation by a heme-containing dioxygenase - PubMed (original) (raw)
Crystal structure of human indoleamine 2,3-dioxygenase: catalytic mechanism of O2 incorporation by a heme-containing dioxygenase
Hiroshi Sugimoto et al. Proc Natl Acad Sci U S A. 2006.
Abstract
Human indoleamine 2,3-dioxygenase (IDO) catalyzes the cleavage of the pyrrol ring of L-Trp and incorporates both atoms of a molecule of oxygen (O2). Here we report on the x-ray crystal structure of human IDO, complexed with the ligand inhibitor 4-phenylimidazole and cyanide. The overall structure of IDO shows two alpha-helical domains with the heme between them. A264 of the flexible loop in the heme distal side is in close proximity to the iron. A mutant analysis shows that none of the polar amino acid residues in the distal heme pocket are essential for activity, suggesting that, unlike the heme-containing monooxygenases (i.e., peroxidase and cytochrome P450), no protein group of IDO is essential in dioxygen activation or proton abstraction. These characteristics of the IDO structure provide support for a reaction mechanism involving the abstraction of a proton from the substrate by iron-bound dioxygen. Inactive mutants (F226A, F227A, and R231A) retain substrate-binding affinity, and an electron density map reveals that 2-(N-cyclohexylamino)ethane sulfonic acid is bound to these residues, mimicking the substrate. These findings suggest that strict shape complementarities between the indole ring of the substrate and the protein side chains are required, not for binding, but, rather, to permit the interaction between the substrate and iron-bound dioxygen in the first step of the reaction. This study provides the structural basis for a heme-containing dioxygenase mechanism, a missing piece in our understanding of heme chemistry.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
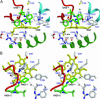
Fig. 1.
Structure of IDO–PI complex. (A) Ribbon representation of the overall structure of human IDO. The small and large domains are represented by blue and green ribbons, respectively. The helices A–S are named in the order of appearance in the primary sequence. The connecting helices (K-L and N) are colored in cyan. The long loop connecting the two domains is colored in red. The heme (yellow), proximal ligand H346 (white), and heme inhibitor 4-phynylimidazole (white) are shown in a ball-and-stick model. The helices of the large domain create the cavity for the heme. The connecting loop (red) and small domain above the sixth-coordination site (heme distal side) cover the top of cavity on the heme. (B) The four proximal helices I, G, Q, and S run in parallel. The helices N (blue) and K-L (cyan) connect the two domains. The connecting loop (red) and small domain above the sixth-coordination site of the heme cover the top of the cavity on the heme.
Fig. 2.
The solvent-accessible surface with an electrostatic potential of IDO. Positive potentials are drawn in blue, negative are in red. The heme is shown as a ball-and-stick model. Unlike as in monooxygenase P450, an asymmetric distribution of positively and negatively charged areas is not observed in IDO. The 7-propionate is partially exposed to the solvent.
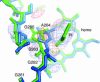
Fig. 3.
Active site of IDO–PI complex. (A) Stereoview of the residues around the heme of IDO viewed from the side of heme plane. The proximal ligand H346 is H-bonded to wa1. The 6-propionate of the heme contacts with wa2 and R343 Nε. The wa2 is H-bonded to wa1, L388 O, and 6-propionate. Mutations of F226, F227, and R231 do not lose the substrate affinity but produce the inactive enzyme. Two CHES molecules are bound in the distal pocket. The cyclohexan ring of CHES-1 (green) contacts with F226 and R231. The 7-propionate of the heme interacts with the amino group of CHES-1 and side chain of Ser-263. The mutational analyses for these distal residues are shown in Table 1. (B) Top view of A by a rotation of 90°. The proximal residues are omitted.
Fig. 4.
Comparison of the PI- and CN−-bound IDO viewed from the distal side. The PI- and CN−-bound structures are shown as stick models in green and blue, respectively. The ligand exchange (from PI to CN−) induces a conformational change in the main chain of the connecting loop. The largest movement is observed in the A264–G265 region with a 1.3-Å shift toward the center of the heme. These models are superimposed on a _F_obs (CN− form) − _F_obs (PI form) difference Fourier map calculated from the phase of PI form. The negative density at −3.0 σ and the positive density at 3.0 σ are shown in light blue and red, respectively.
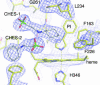
Fig. 5.
The electron density of a 2 _F_obs − _F_calc simulated annealing composite omit map (generated with 5% of the overall model omitted) around the heme of PI form at 2.3-Å resolution is contoured at 1.2 σ. The final refined model is superimposed.
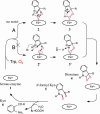
Fig. 6.
The proposed catalytic mechanism. IDO catalyzes the cleavage of the bond between 2-C and 3-C of its substrate
l
-Trp. The trigger for the reaction must be the abstraction of a proton from 1-N. There have been two possible pathways after the formation of ternary complex. Terentis et al. (17) proposed a pathway that involves proton abstraction by iron-bound dioxygen. Their model is modified based on the tertiary structure in our proposed model (scheme A). The binding of O2 and the substrate
l
-Trp, whose orientation is restricted by F226 and R231, enables an interaction between the NH group of indole and the proximal atom of dioxygen (2). The proton is then abstracted from 1-N by dioxygen. The rearrangement of the electronic structure of the indole ring induces an electrophilic reaction, which involves the formation of a bond between the terminal oxygen atom of dioxygen and the 3-C atom. The subsequent cleavage of the Fe O bond results in the formation of the 3-hydroperoxyindolenine intermediate (3). In scheme B, a protein base abstracts the proton of 1-NH. The dioxetane (4) has been proposed as the intermediate during the incorporation of dioxygen. The product _N_-formyl Kyn (5) is converted to Kyn (6) nonenzymatically or by Kyn formamidase (33).
O bond results in the formation of the 3-hydroperoxyindolenine intermediate (3). In scheme B, a protein base abstracts the proton of 1-NH. The dioxetane (4) has been proposed as the intermediate during the incorporation of dioxygen. The product _N_-formyl Kyn (5) is converted to Kyn (6) nonenzymatically or by Kyn formamidase (33).
Similar articles
- Density functional theory study on a missing piece in understanding of heme chemistry: the reaction mechanism for indoleamine 2,3-dioxygenase and tryptophan 2,3-dioxygenase.
Chung LW, Li X, Sugimoto H, Shiro Y, Morokuma K. Chung LW, et al. J Am Chem Soc. 2008 Sep 17;130(37):12299-309. doi: 10.1021/ja803107w. Epub 2008 Aug 20. J Am Chem Soc. 2008. PMID: 18712870 - UV Resonance Raman Characterization of a Substrate Bound to Human Indoleamine 2,3-Dioxygenase 1.
Yanagisawa S, Kayama K, Hara M, Sugimoto H, Shiro Y, Ogura T. Yanagisawa S, et al. Biophys J. 2019 Aug 20;117(4):706-716. doi: 10.1016/j.bpj.2019.07.017. Epub 2019 Jul 19. Biophys J. 2019. PMID: 31405517 Free PMC article. - ONIOM study on a missing piece in our understanding of heme chemistry: bacterial tryptophan 2,3-dioxygenase with dual oxidants.
Chung LW, Li X, Sugimoto H, Shiro Y, Morokuma K. Chung LW, et al. J Am Chem Soc. 2010 Sep 1;132(34):11993-2005. doi: 10.1021/ja103530v. J Am Chem Soc. 2010. PMID: 20698527 - Biochemical properties of indoleamine 2,3-dioxygenase: from structure to optimized design of inhibitors.
Lancellotti S, Novarese L, De Cristofaro R. Lancellotti S, et al. Curr Med Chem. 2011;18(15):2205-14. doi: 10.2174/092986711795656108. Curr Med Chem. 2011. PMID: 21517759 Review. - Highlights at the gate of tryptophan catabolism: a review on the mechanisms of activation and regulation of indoleamine 2,3-dioxygenase (IDO), a novel target in cancer disease.
Macchiarulo A, Camaioni E, Nuti R, Pellicciari R. Macchiarulo A, et al. Amino Acids. 2009 Jul;37(2):219-29. doi: 10.1007/s00726-008-0137-3. Epub 2008 Jul 9. Amino Acids. 2009. PMID: 18612775 Review.
Cited by
- Docking Studies and Molecular Dynamic Simulations Reveal Different Features of IDO1 Structure.
Greco FA, Bournique A, Coletti A, Custodi C, Dolciami D, Carotti A, Macchiarulo A. Greco FA, et al. Mol Inform. 2016 Sep;35(8-9):449-59. doi: 10.1002/minf.201501038. Epub 2016 Jul 19. Mol Inform. 2016. PMID: 27546049 Free PMC article. - Inhibitory substrate binding site of human indoleamine 2,3-dioxygenase.
Lu C, Lin Y, Yeh SR. Lu C, et al. J Am Chem Soc. 2009 Sep 16;131(36):12866-7. doi: 10.1021/ja9029768. J Am Chem Soc. 2009. PMID: 19737010 Free PMC article. - Substrate and cofactor requirements of indoleamine 2,3-dioxygenase in interferon-gamma-treated cells: utilization of oxygen rather than superoxide.
Werner ER, Werner-Felmayer G. Werner ER, et al. Curr Drug Metab. 2007 Apr;8(3):201-3. doi: 10.2174/138920007780362482. Curr Drug Metab. 2007. PMID: 17430107 Free PMC article. Review. - Ferryl derivatives of human indoleamine 2,3-dioxygenase.
Lu C, Yeh SR. Lu C, et al. J Biol Chem. 2011 Jun 17;286(24):21220-30. doi: 10.1074/jbc.M111.221507. Epub 2011 Apr 18. J Biol Chem. 2011. PMID: 21502325 Free PMC article.
References
- Hayaishi O., Nozaki M. Science. 1969;164:389–396. - PubMed
- Yamamoto S., Hayaishi O. J. Biol. Chem. 1967;242:5260–5266. - PubMed
- Hayaishi O., Rothberg S., Mehler A. H., Saito Y. J. Biol. Chem. 1957;229:889–896. - PubMed
- Knox W. E., Mehler A. H. J. Biol. Chem. 1950;187:419–430. - PubMed
- Takikawa O., Truscott R. J., Fukao M., Miwa S. Adv. Exp. Med. Biol. 2003;527:277–285. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials