High-throughput trapping of secretory pathway genes in mouse embryonic stem cells - PubMed (original) (raw)
High-throughput trapping of secretory pathway genes in mouse embryonic stem cells
Silke De-Zolt et al. Nucleic Acids Res. 2006.
Abstract
High-throughput gene trapping is a random approach for inducing insertional mutations across the mouse genome. This approach uses gene trap vectors that simultaneously inactivate and report the expression of the trapped gene at the insertion site, and provide a DNA tag for the rapid identification of the disrupted gene. Gene trapping has been used by both public and private institutions to produce libraries of embryonic stem (ES) cells harboring mutations in single genes. Presently, approximately 66% of the protein coding genes in the mouse genome have been disrupted by gene trap insertions. Among these, however, genes encoding signal peptides or transmembrane domains (secretory genes) are underrepresented because they are not susceptible to conventional trapping methods. Here, we describe a high-throughput gene trapping strategy that effectively targets secretory genes. We used this strategy to assemble a library of ES cells harboring mutations in 716 unique secretory genes, of which 61% were not trapped by conventional trapping, indicating that the two strategies are complementary. The trapped ES cell lines, which can be ordered from the International Gene Trap Consortium (http://www.genetrap.org), are freely available to the scientific community.
Figures
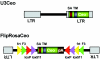
**Figure 1
Schematic representation of the secretory gene trap vectors. LTR, long terminal repeat; Ceo, CD2/neomycinphosphotransferase fusion gene; SA, splice acceptor; TM, transmembrane domain; frt (yellow triangles) and F3 (green triangles) heterotypic target sequences for FLPe recombinase; loxP (red triangle) and lox511 (purple triangles), heterotypic target sequences from Cre recombinase.
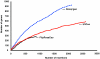
**Figure 2
Comparison of the rates of trapping of secretory and standard gene trap vectors. GTSTs matching unique genes (e ≤ 10−6) were identified by using the RefSeq database (rel. 13) and the BlastN algorithm. The number of novel genes hit by accumulating insertions was traced chronologically.
**Figure 3
Correlation between secretory trap insertions and the number of individual genes per chromosome.
**Figure 4
Distribution of gene trap insertions according to the position of the trapped intron within genes. The data are based on NCBI mouse genome build 34 and RefSeq database release 13.
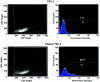
**Figure 5
CD2 expression by U3Ceo trapped ES cells. A total of 5 × 104 cells were infected with U3Ceo retrovirus selected in G418. A total of 1 × 106 NeoR cells (trapped TBV-2) were treated with the Leu-5b monoclonal mouse anti-human CD2 antibody. Similarly treated wild-type (TBV-2) cells served as a negative control. CD2 expression was estimated by flow cytometry after treating the cells with a FITC-conjugated sheep anti-mouse Fab′2 antibody.
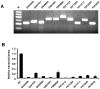
**Figure 6
Transcriptional analysis of X-linked mutations in trapped ES cell lines. (A) RT–PCR of fusion transcripts induced by the gene trap insertions using trapped gene- and vector- specific primers. M, molecular weight marker (50 bp ladder). (B) Quantitative RT–PCR of wild-type transcripts expressed in the trapped ES cell lines. Gene-specific primers were chosen in exons flanking the insertion sites (Supplementary Table 1 and Supplementary Figure 1). Results are means from three independent experiments ± SD.
**Figure 7
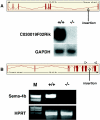
Secretory gene disruption in transgenic mice. (A) Top: structure of the C030019F02Rik gene as annotated by ENSEMBL and position of the gene trap insertions (arrow). Bottom: northern blot analysis of C030019F02Rik transcripts expressed in homozygous mice. 20 µg total RNAs extracted from the brains of wild type (+/+) and mutant (−/−) mice were fractionated on 1% formaldehyde/agarose gels and hybridized to a 32P-labeled full-length C030019F02Rik c-DNA probe. (B) Top: structure of the semaphorin-4B gene as annotated by ENSEMBL and position of the gene trap insertion (arrow). Bottom: RT–PCR of transcripts expressed in wild type (+/+) and homozygous mutant (−/−) embryos using primers complementary to the 11th and 14th exon of the semaphorin-4B gene. M, molecular weight marker (smart ladder).
Similar articles
- Suppression of nonsense-mediated mRNA decay permits unbiased gene trapping in mouse embryonic stem cells.
Shigeoka T, Kawaichi M, Ishida Y. Shigeoka T, et al. Nucleic Acids Res. 2005 Feb 1;33(2):e20. doi: 10.1093/nar/gni022. Nucleic Acids Res. 2005. PMID: 15687378 Free PMC article. - Genomewide production of multipurpose alleles for the functional analysis of the mouse genome.
Schnütgen F, De-Zolt S, Van Sloun P, Hollatz M, Floss T, Hansen J, Altschmied J, Seisenberger C, Ghyselinck NB, Ruiz P, Chambon P, Wurst W, von Melchner H. Schnütgen F, et al. Proc Natl Acad Sci U S A. 2005 May 17;102(20):7221-6. doi: 10.1073/pnas.0502273102. Epub 2005 May 3. Proc Natl Acad Sci U S A. 2005. PMID: 15870191 Free PMC article. - Gene-trap vectors and mutagenesis.
De-Zolt S, Altschmied J, Ruiz P, von Melchner H, Schnütgen F. De-Zolt S, et al. Methods Mol Biol. 2009;530:29-47. doi: 10.1007/978-1-59745-471-1_3. Methods Mol Biol. 2009. PMID: 19266330 - The gene trap resource: a treasure trove for hemopoiesis research.
Forrai A, Robb L. Forrai A, et al. Exp Hematol. 2005 Aug;33(8):845-56. doi: 10.1016/j.exphem.2005.03.016. Exp Hematol. 2005. PMID: 16038776 Review. - A review of current large-scale mouse knockout efforts.
Guan C, Ye C, Yang X, Gao J. Guan C, et al. Genesis. 2010 Feb;48(2):73-85. doi: 10.1002/dvg.20594. Genesis. 2010. PMID: 20095055 Review.
Cited by
- Mutant non-coding RNA resource in mouse embryonic stem cells.
Hansen J, von Melchner H, Wurst W. Hansen J, et al. Dis Model Mech. 2021 Feb 5;14(2):dmm047803. doi: 10.1242/dmm.047803. Dis Model Mech. 2021. PMID: 33729986 Free PMC article. - Gene trap mutagenesis in the mouse.
Friedel RH, Soriano P. Friedel RH, et al. Methods Enzymol. 2010;477:243-69. doi: 10.1016/S0076-6879(10)77013-0. Methods Enzymol. 2010. PMID: 20699145 Free PMC article. - Gene-trapped mouse embryonic stem cell-derived cardiac myocytes and human genetics implicate AKAP10 in heart rhythm regulation.
Tingley WG, Pawlikowska L, Zaroff JG, Kim T, Nguyen T, Young SG, Vranizan K, Kwok PY, Whooley MA, Conklin BR. Tingley WG, et al. Proc Natl Acad Sci U S A. 2007 May 15;104(20):8461-6. doi: 10.1073/pnas.0610393104. Epub 2007 May 7. Proc Natl Acad Sci U S A. 2007. PMID: 17485678 Free PMC article. - The carnegie protein trap library: a versatile tool for Drosophila developmental studies.
Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, Skora AD, Nystul TG, Ohlstein B, Allen A, Wilhelm JE, Murphy TD, Levis RW, Matunis E, Srivali N, Hoskins RA, Spradling AC. Buszczak M, et al. Genetics. 2007 Mar;175(3):1505-31. doi: 10.1534/genetics.106.065961. Epub 2006 Dec 28. Genetics. 2007. PMID: 17194782 Free PMC article. - PiggyBac transposon-based polyadenylation-signal trap for genome-wide mutagenesis in mice.
Li L, Liu P, Sun L, Bin Zhou, Fei J. Li L, et al. Sci Rep. 2016 Jun 13;6:27788. doi: 10.1038/srep27788. Sci Rep. 2016. PMID: 27292714 Free PMC article.
References
- Stanford W.L., Cohn J.B., Cordes S.P. Gene-trap mutagenesis: past, present and beyond. Nature Rev. Genet. 2001;2:756–768. - PubMed
- Zambrowicz B.P., Abuin A., Ramirez-Solis R., Richter L.J., Piggott J., BeltrandelRio H., Buxton E.C., Edwards J., Finch R.A., Friddle C.J., et al. Wnk1 kinase deficiency lowers blood pressure in mice: A gene-trap screen to identify potential targets for therapeutic intervention. Proc. Natl Acad. Sci. USA. 2003;100:14109–14114. - PMC - PubMed
- Wiles M.V., Vauti F., Otte J., Fuchtbauer E.M., Ruiz P., Fuchtbauer A., Arnold H.H., Lehrach H., Metz T., von Melchner H., et al. Establishment of a gene-trap sequence tag library to generate mutant mice from embryonic stem cells. Nature Genet. 2000;24:13–14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases