Active role for nibrin in the kinetics of atm activation - PubMed (original) (raw)
Active role for nibrin in the kinetics of atm activation
Karen Cerosaletti et al. Mol Cell Biol. 2006 Mar.
Abstract
The Atm protein kinase is central to the DNA double-strand break response in mammalian cells. After irradiation, dimeric Atm undergoes autophosphorylation at Ser 1981 and dissociates into active monomers. Atm activation is stimulated by expression of the Mre11/Rad50/nibrin complex. Previously, we showed that a C-terminal fragment of nibrin, containing binding sites for both Mre11 and Atm, was sufficient to provide this stimulatory effect in Nijmegen breakage syndrome (NBS) cells. To discriminate whether nibrin's role in Atm activation is to bind and translocate Mre11/Rad50 to the nucleus or to interact directly with Atm, we expressed an Mre11 transgene with a C-terminal NLS sequence in NBS fibroblasts. The Mre11-NLS protein complexed with Rad50, localized to the nucleus in NBS fibroblasts, and associated with chromatin. However, Atm autophosphorylation was not stimulated in cells expressing Mre11-NLS, nor were downstream Atm targets phosphorylated. To determine whether nibrin-Atm interaction is necessary to stimulate Atm activation, we expressed nibrin transgenes lacking the Atm binding domain in NBS fibroblasts. The nibrin DeltaAtm protein interacted with Mre11/Rad50; however, Atm autophosphorylation was dramatically reduced after irradiation in NBS cells expressing the nibrin DeltaAtm transgenes relative to wild-type nibrin. These results indicate that nibrin plays an active role in Atm activation beyond translocating Mre11/Rad50 to the nucleus and that this function requires nibrin-Atm interaction.
Figures
FIG. 1.
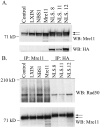
Expression of the Mre11-NLS transgene in NBS cells and interaction with Rad50. (A) Western blot analysis of GM0637 fibroblasts or NBS-ILB1 cells stably transduced with the pLXIN retroviral vector alone (LXIN), wild-type nibrin (NBS1), wild-type Mre11 (Mre11), or an Mre11-NLS transgene with two artificial NLS sequences and an HA tag attached to the Mre11 C terminus. Individual clones of NBS cells expressing the Mre11-NLS transgene were isolated (NLS.8, NLS.11, and NLS.12). A total of 25 μg of total cellular protein per lane was separated on a 7% sodium dodecyl sulfate (SDS)-polyacrylamide gel. The Western blot (WB) was probed with monoclonal antibodies specific for Mre11 or the HA tag. (B) Mre11 was immunoprecipitated (IP) from 250 μg of total cellular protein isolated from the above cell lines using a polyclonal anti-Mre11 antibody or a monoclonal anti-HA antibody. Immunoprecipitated proteins were separated on a 3 to 8% SDS-polyacrylamide gel, and the immunoblot was probed with a polyclonal Rad50 antisera or a monoclonal anti-Mre11 antibody. The arrows indicate the positions of endogenous Mre11 protein and the higher-molecular-weight Mre11-NLS protein.
FIG. 2.
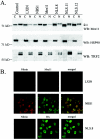
Subcellular localization of the Mre11-NLS protein in NBS cells. (A) Cytoplasmic and nuclear proteins were isolated from GM0637 control fibroblasts or NBS-ILB1 cells stably transduced with the pLXIN retroviral vector alone (LXIN), wild-type nibrin (NBS1), wild-type Mre11 (Mre11), or the Mre11-NLS transgene (NLS.8, NLS.11, and NLS.12). A total of 105 cell equivalents of protein per lane were electrophoresed on 7% SDS-polyacrylamide gels. Western blots (WB) were probed with a monoclonal anti-Mre11 antibody, a polyclonal antisera specific for the cytoplasmic heat shock protein (Hsp90), or a monoclonal antibody specific for the telomeric protein, TRF2. The arrows indicate the positions of endogenous Mre11 protein and the higher-molecular-weight Mre11-NLS protein. (B) Immunofluorescence analysis was performed on NBS-ILB1 cells stably transduced with the pLXIN retroviral vector alone, wild-type nibrin (NBS1), or the Mre11-NLS transgene (NLS.8). Fibroblasts were stained for nibrin and Mre11 expression using a polyclonal anti-nibrin antibody (red) and a monoclonal antibody to Mre11 (green). The Mre11-NLS protein was specifically detected using a monoclonal antibody specific for the HA tag (HA, green). Immunofluorescence was detected by confocal microscopy at 488 and 568 nm. Individual fields were Z-planed and stacked. Magnification, ×600.
FIG. 3.
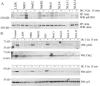
Atm activation and function after irradiation in NBS cells expressing the Mre11-NLS protein. (A) Atm autophosphorylation at serine 1981 was assessed in NBS-ILB1 cells stably transduced with the pLXIN vector alone (LXIN), wild-type nibrin (NBS1), nibrin lacking the C-terminal 100 aa (Nb652), a C-terminal 300-aa fragment of nibrin (NbFR5), wild-type Mre11 (Mre11), or the Mre11-NLS transgene (NLS.8, NLS.11, and NLS.12). Cells were unirradiated or exposed to 3 Gy of ionizing radiation (IR), and total cellular lysates were harvested after 15 min. Atm was immunoprecipitated (IP) from 800 μg of protein using a polyclonal anti-Atm antibody and protein was separated on 3 to 8% SDS-polyacrylamide gels. Western blots (WB) were probed with a monoclonal phosphoserine 1981 antibody or a polyclonal Atm antibody. (B) Chk2 threonine 68 phosphorylation and p53 serine 15 phosphorylation by Atm were assessed in the above cells, as well as GM0637 control fibroblasts, 15 min after exposure to 0 or 3 Gy of ionizing radiation. A total of 20 μg of total cellular protein per lane was separated on 7% SDS-polyacrylamide gels. Immunoblots were probed with a phosphothreonine 68 antisera or a monoclonal anti-Chk2 antibody. For p53, the immunoblots were probed with phosphoserine 15 antisera or a monoclonal p53 antibody.
FIG. 4.
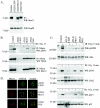
Atm activation and function after irradiation in ATLD cells expressing the Mre11-NLS protein. (A) Expression of the Mre11 NLS transgene in ATLD cells was assessed by Western blot analysis. Total cellular lysates were isolated from GM0637 control fibroblasts and ATLD D6809 cells stably transduced with the pLXIN retroviral vector (ATLD LXIN), wild-type Mre11 (ATLD Mre11), or the Mre11-NLS transgene (ATLD NLS). Then, 25 μg of protein per lane was electrophoresed on a 7% SDS-polyacrylamide gel and Western blotted (WB). The immunoblot was probed with a monoclonal anti-Mre11 antibody or a polyclonal antibody specific for Hsp90, as a loading control. The arrows indicate the endogenous Mre11 protein and the higher-molecular-weight Mre11-NLS protein. (B) Nibrin phosphorylation in the above cell lines was analyzed 15 min after exposure to 0 or 3 Gy of ionizing radiation (IR). Nibrin was immunoprecipitated (IP) from 250 μg of total cellular protein using a polyclonal antinibrin antibody and proteins were separated on a 3 to 8% SDS-polyacrylamide gel. Western blots were probed with a phosphoserine 343 monoclonal antibody or a polyclonal antibody specific for nibrin. To assess Mre11-NLS complex formation with nibrin, the blot was reprobed with a monoclonal anti-Mre11 antibody. (C) Atm autophosphorylation at serine 1981 was assessed in the above cells lines 15 min after exposure to 0 or 3 Gy of irradiation. Next, 25 μg of total cellular protein per lane was electrophoresed on a 3 to 8% SDS-polyacrylamide gel. The Western blot was probed with a monoclonal phosphoserine 1981 antibody or a polyclonal anti-Atm antisera. Chk2 threonine 68 phosphorylation was analyzed by probing a similar immunoblot with phosphothreonine 68 antisera or a monoclonal anti-Chk2 antibody. To detect p53 serine 15 phosphorylation, immunoblots were probed with phosphoserine 15 antisera or a monoclonal p53 antibody. (D) Irradiation-induced foci were examined 6 h after 12 Gy of irradiation in the above ATLD fibroblasts. Mre11 or Mre11-NLS foci were detected with Mre11 antisera (red) and γ-H2AX foci were detected with a monoclonal γ-H2AX antibody (green). Immunofluorescence was collected by using confocal microscopy at 488 and 568 nm. Individual fields were Z-planed and stacked. Magnification, ×1,000.
FIG. 5.
Localization of the Mre11-NLS protein to chromatin in NBS cells. NBS-ILB1 cells stably transduced with the pLXIN retroviral vector alone (LXIN), wild-type nibrin (NBS1), or the Mre11-NLS transgene (Mre11-NLS) were unirradiated or exposed to 3 Gy of ionizing radiation. After 30 min cells were harvested and fractionated into cytoplasmic (lanes C), nuclear soluble (lanes NS), nuclear 125 mM NaCl wash (lanes 125), nuclear 250 mM NaCl wash (lanes 250), and the insoluble nuclear pellet (lanes NP). For comparison, whole-cell lysate was included (lanes WC). A total of 105 cell equivalents of protein per lane was separated on 3 to 8% SDS-polyacrylamide gels and Western blotted (WB). (A) Immunoblots of pLXIN cellular fractions were probed with an anti-Mre11 monoclonal antibody to assess the localization of Mre11 in the absence of full-length nibrin. The blot was reprobed with anti-Hsp90 antisera as a control for cytoplasmic proteins or with a monoclonal TRF2 antibody as a control for nuclear proteins. (B) Localization of Mre11 in cellular fractions from LXIN, NBS1, and Mre11-NLS expressing cells was detected by probing immunoblots with a monoclonal Mre11 antibody.
FIG. 6.
Expression of nibrin ΔAtm transgenes in NBS cells and Atm activation after irradiation. (A) Expression and MRN complex formation was analyzed in GM0637 control fibroblasts and NBS-ILB1 cells stably transduced with the pLXIN retroviral vector alone (LXIN), wild-type nibrin (NBS1), nibrin lacking the C-terminal Atm binding domain (NBS1 ΔAtm), a 300-aa C-terminal fragment of nibrin (NbFR5), and NbFR5 lacking the C-terminal Atm binding domain (NbFR5 ΔAtm). Nibrin was immunoprecipitated (IP) from 250 μg of total cellular protein using polyclonal nibrin antisera and proteins were separated on a 3 to 8% SDS-polyacrylamide gel. The Western blot (WB) was probed with an anti-Mre11 monoclonal antibody, a polyclonal Rad50 antibody, or a monoclonal nibrin antibody. (B) Atm autophosphorylation at serine 1981 was assessed in the above cells 15 min after treatment with 0 or 3 Gy of ionizing radiation (IR). Atm was immunoprecipitated from 800 μg of total cellular protein using a polyclonal Atm antibody and proteins were separated on 3 to 8% SDS-polyacrylamide gels. Immunoblots were probed with a monoclonal phosphoserine 1981 antibody or a polyclonal Atm antibody. To detect Chk2 and p53 phosphorylation, 20 μg of total cellular protein isolated from the above cells 15 min after 0 or 3 Gy of irradiation was separated on 7% SDS-polyacrylamide gels. Immunoblots were probed with phosphothreonine 68 antisera or a Chk2 monoclonal antibody. For p53, Western blots were probed with phosphoserine 15 antisera or a monoclonal p53 antibody.
Similar articles
- Independent roles for nibrin and Mre11-Rad50 in the activation and function of Atm.
Cerosaletti K, Concannon P. Cerosaletti K, et al. J Biol Chem. 2004 Sep 10;279(37):38813-9. doi: 10.1074/jbc.M404294200. Epub 2004 Jul 1. J Biol Chem. 2004. PMID: 15234984 - Cleavage of the BRCT tandem domains of nibrin by the 657del5 mutation affects the DNA damage response less than the Arg215Trp mutation.
Mendez G, Cilli D, Berardinelli F, Viganotti M, Ascenzi P, Tanzarella C, Antoccia A, di Masi A. Mendez G, et al. IUBMB Life. 2012 Oct;64(10):853-61. doi: 10.1002/iub.1077. Epub 2012 Sep 3. IUBMB Life. 2012. PMID: 22941933 - Ataxia telangiectasia-mutated (ATM) kinase activity is regulated by ATP-driven conformational changes in the Mre11/Rad50/Nbs1 (MRN) complex.
Lee JH, Mand MR, Deshpande RA, Kinoshita E, Yang SH, Wyman C, Paull TT. Lee JH, et al. J Biol Chem. 2013 May 3;288(18):12840-51. doi: 10.1074/jbc.M113.460378. Epub 2013 Mar 22. J Biol Chem. 2013. PMID: 23525106 Free PMC article. - Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template.
Williams RS, Williams JS, Tainer JA. Williams RS, et al. Biochem Cell Biol. 2007 Aug;85(4):509-20. doi: 10.1139/O07-069. Biochem Cell Biol. 2007. PMID: 17713585 Review. - Activation and regulation of ATM kinase activity in response to DNA double-strand breaks.
Lee JH, Paull TT. Lee JH, et al. Oncogene. 2007 Dec 10;26(56):7741-8. doi: 10.1038/sj.onc.1210872. Oncogene. 2007. PMID: 18066086 Review.
Cited by
- A systematic proteomic study of irradiated DNA repair deficient Nbn-mice.
Melchers A, Stöckl L, Radszewski J, Anders M, Krenzlin H, Kalischke C, Scholz R, Jordan A, Nebrich G, Klose J, Sperling K, Digweed M, Demuth I. Melchers A, et al. PLoS One. 2009;4(5):e5423. doi: 10.1371/journal.pone.0005423. Epub 2009 May 1. PLoS One. 2009. PMID: 19412544 Free PMC article. - The Drosophila Nbs protein functions in multiple pathways for the maintenance of genome stability.
Ciapponi L, Cenci G, Gatti M. Ciapponi L, et al. Genetics. 2006 Jul;173(3):1447-54. doi: 10.1534/genetics.106.058081. Epub 2006 Apr 30. Genetics. 2006. PMID: 16648644 Free PMC article. - Replication independent ATR signalling leads to G2/M arrest requiring Nbs1, 53BP1 and MDC1.
Stiff T, Cerosaletti K, Concannon P, O'Driscoll M, Jeggo PA. Stiff T, et al. Hum Mol Genet. 2008 Oct 15;17(20):3247-53. doi: 10.1093/hmg/ddn220. Epub 2008 Jul 28. Hum Mol Genet. 2008. PMID: 18664457 Free PMC article. - Multiple autophosphorylation sites are dispensable for murine ATM activation in vivo.
Daniel JA, Pellegrini M, Lee JH, Paull TT, Feigenbaum L, Nussenzweig A. Daniel JA, et al. J Cell Biol. 2008 Dec 1;183(5):777-83. doi: 10.1083/jcb.200805154. J Cell Biol. 2008. PMID: 19047460 Free PMC article. - Tip60: connecting chromatin to DNA damage signaling.
Sun Y, Jiang X, Price BD. Sun Y, et al. Cell Cycle. 2010 Mar 1;9(5):930-6. doi: 10.4161/cc.9.5.10931. Epub 2010 Mar 11. Cell Cycle. 2010. PMID: 20160506 Free PMC article. Review.
References
- Bakkenist, C. J., and M. B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421:499-506. - PubMed
- Banin, S., L. Moyal, S. Shieh, Y. Taya, C. W. Anderson, L. Chessa, N. I. Smorodinsky, C. Prives, Y. Reiss, Y. Shiloh, and Y. Ziv. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281:1674-1677. - PubMed
- Carney, J. P., R. S. Maser, H. Olivares, E. M. Davis, M. Le Beau, J. R. Yates III, L. Hays, W. F. Morgan, and J. H. Petrini. 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93:477-486. - PubMed
- Cerosaletti, K., and P. Concannon. 2004. Independent roles for nibrin and Mre11-Rad50 in the activation and function of Atm. J. Biol. Chem. 279:38813-38819. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous