N6-methyladenine: the other methylated base of DNA - PubMed (original) (raw)
Review
N6-methyladenine: the other methylated base of DNA
David Ratel et al. Bioessays. 2006 Mar.
Abstract
Contrary to mammalian DNA, which is thought to contain only 5-methylcytosine (m5C), bacterial DNA contains two additional methylated bases, namely N6-methyladenine (m6A), and N4-methylcytosine (m4C). However, if the main function of m5C and m4C in bacteria is protection against restriction enzymes, the roles of m6A are multiple and include, for example, the regulation of virulence and the control of many bacterial DNA functions such as the replication, repair, expression and transposition of DNA. Interestingly, even if adenine methylation is usually considered a bacterial DNA feature, the presence of m6A has been found in protist and plant DNAs. Furthermore, indirect evidence suggests the presence of m6A in mammal DNA, raising the possibility that this base has remained undetected due to the low sensitivity of the analytical methods used. This highlights the importance of considering m6A as the sixth element of DNA.
Copyright 2006 Wiley Periodicals, Inc.
Figures
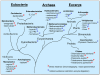
Fig. 1
Schematic representation of the phylogenetic distribution of m6A in DNA.
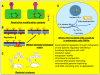
Fig. 2. Some established and putative functions of m6A in DNA
A) Restriction-modifications systems defend bacteria from invasions by viral DNA. These systems are composed of a bacterial restriction enzyme which cuts a specific sequence in phage DNA, and a cognate adenine or cytosine DNA methyltransferase which recognizes the same target site that the restriction enzyme and protects bacterial DNA from its own restriction enzyme(s). B) In E. coli, adenine methylation is also used to discriminate the transiently unmethylated newly synthesized complementary strand during DNA replication. This discrimination is the basis of the methyl-directed mismatch repair which makes correction on the newly synthesized strand only. C) Alteration in the levels of DNA adenine methylation attenuates the virulence of a number of pathogens. D) Speculative representation of the biological functions of putative eucaryotic N-6 adenine-specific DNA methylase(s).
Similar articles
- DNA methylation in thermophilic bacteria: N4-methylcytosine, 5-methylcytosine, and N6-methyladenine.
Ehrlich M, Gama-Sosa MA, Carreira LH, Ljungdahl LG, Kuo KC, Gehrke CW. Ehrlich M, et al. Nucleic Acids Res. 1985 Feb 25;13(4):1399-412. doi: 10.1093/nar/13.4.1399. Nucleic Acids Res. 1985. PMID: 4000939 Free PMC article. - Undetectable levels of N6-methyl adenine in mouse DNA: Cloning and analysis of PRED28, a gene coding for a putative mammalian DNA adenine methyltransferase.
Ratel D, Ravanat JL, Charles MP, Platet N, Breuillaud L, Lunardi J, Berger F, Wion D. Ratel D, et al. FEBS Lett. 2006 May 29;580(13):3179-84. doi: 10.1016/j.febslet.2006.04.074. Epub 2006 May 3. FEBS Lett. 2006. PMID: 16684535 - An Adenine Code for DNA: A Second Life for N6-Methyladenine.
Heyn H, Esteller M. Heyn H, et al. Cell. 2015 May 7;161(4):710-3. doi: 10.1016/j.cell.2015.04.021. Epub 2015 Apr 30. Cell. 2015. PMID: 25936836 Review. - Detection of N6-methyladenine in GATC sequences of Selenomonas ruminantium.
Pristas P, Molnarova V, Javorsky P. Pristas P, et al. J Basic Microbiol. 1998;38(4):283-7. J Basic Microbiol. 1998. PMID: 9791949 - Role of DNA and RNA N6-Adenine Methylation in Regulating Stem Cell Fate.
Wu Y, Zhou C, Yuan Q. Wu Y, et al. Curr Stem Cell Res Ther. 2018;13(1):31-38. doi: 10.2174/1574888X12666170621125457. Curr Stem Cell Res Ther. 2018. PMID: 28637404 Review.
Cited by
- Abundant DNA 6mA methylation during early embryogenesis of zebrafish and pig.
Liu J, Zhu Y, Luo GZ, Wang X, Yue Y, Wang X, Zong X, Chen K, Yin H, Fu Y, Han D, Wang Y, Chen D, He C. Liu J, et al. Nat Commun. 2016 Oct 7;7:13052. doi: 10.1038/ncomms13052. Nat Commun. 2016. PMID: 27713410 Free PMC article. - Production and characterization of novel recombinant adeno-associated virus replicative-form genomes: a eukaryotic source of DNA for gene transfer.
Li L, Dimitriadis EK, Yang Y, Li J, Yuan Z, Qiao C, Beley C, Smith RH, Garcia L, Kotin RM. Li L, et al. PLoS One. 2013 Aug 1;8(8):e69879. doi: 10.1371/journal.pone.0069879. Print 2013. PLoS One. 2013. PMID: 23936358 Free PMC article. - Nucleic acid oxidation in DNA damage repair and epigenetics.
Zheng G, Fu Y, He C. Zheng G, et al. Chem Rev. 2014 Apr 23;114(8):4602-20. doi: 10.1021/cr400432d. Epub 2014 Feb 28. Chem Rev. 2014. PMID: 24580634 Free PMC article. Review. No abstract available. - DCNN-4mC: Densely connected neural network based N4-methylcytosine site prediction in multiple species.
Rehman MU, Tayara H, Chong KT. Rehman MU, et al. Comput Struct Biotechnol J. 2021 Nov 1;19:6009-6019. doi: 10.1016/j.csbj.2021.10.034. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34849205 Free PMC article. - MASQC: Next Generation Sequencing Assists Third Generation Sequencing for Quality Control in N6-Methyladenine DNA Identification.
Yang S, Wang Y, Chen Y, Dai Q. Yang S, et al. Front Genet. 2020 Mar 24;11:269. doi: 10.3389/fgene.2020.00269. eCollection 2020. Front Genet. 2020. PMID: 32269589 Free PMC article.
References
- Doskocil J, Sormo’Va Z. Biochim Biophys Acta. 1965 Mar 15;95:513–5. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources