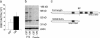Disc1 is mutated in the 129S6/SvEv strain and modulates working memory in mice - PubMed (original) (raw)
Disc1 is mutated in the 129S6/SvEv strain and modulates working memory in mice
Hiroko Koike et al. Proc Natl Acad Sci U S A. 2006.
Abstract
Disrupted-In-Schizophrenia (DISC1) is a leading candidate schizophrenia susceptibility gene. Here, we describe a deletion variant in mDisc1 specific to the 129S6/SvEv strain that introduces a termination codon at exon 7, abolishes production of the full-length protein, and impairs working memory performance when transferred to the C57BL/6J genetic background. Our findings provide insights into how DISC1 variation contributes to schizophrenia susceptibility in humans and the behavioral divergence between 129S6/SvEv and C57BL/6J mouse strains and have implications for modeling psychiatric diseases in mice.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
Fig. 1.
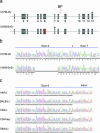
mDisc1 deletion variant in the 129S6/SvEv strain. (a) Genomic structure of mDisc1 gene. The position of the 129S6/SvEv deletion polymorphism in exon 6 is indicated by a red bar. BP, breakpoint in the Scottish family; ∗, premature stop codon. (b) Chromatograms from sequencing mDisc1 exon 6 in C57BL/6J and 129S6/SvEv mouse strains. The 129S6/SvEv 25-bp deletion variant in exon 6 (black horizontal line) and the introduced premature termination codon (∗) at exon 7 are indicated. Exon 6 was amplified from brain cDNA for the 129S6/SvEv and C57BL/6J strains although similar results were obtained from genomic PCR amplification (data not shown). Red arrowhead indicates the splice junction between exons 6 and 7. (c) Chromatograms from sequencing mDisc1 exon 6 in five other mouse strains. Indicated is genomic sequence from mDisc1 exon 6 and surrounding intron 6 in AKR/J, BALB/cJ, CBA/J, C3H/HeJ, and DBA/2J strains.
Fig. 2.
mDisc1 protein levels in the 129S6/SvEv strain. (a) Stability and abundance of the full-length transcript in the 129S6/SvEv strain. Transcript levels were estimated by quantitative RT-PCR on total RNA isolated from P10 mouse brain, by using a C-terminal probe. No internal promoter usage has been described at this locus, and therefore this method provides a good indication of the stability and abundance of the full-length transcript in the 129S6/SvEv strain. C57, C57BL/6J; 129, 129S6/SvEv. (b) The effect of the 129S6/SvEv exon 6 variant on protein expression levels. Shown is Western blot analysis with an antibody raised against the N-terminal region of mDisc1. Three major bands are observed. The ≈96-kDa band most likely corresponds to the predicted 92-kDa full-length protein. The ≈45-kDa signal may correspond to a previously described short form variant (NCBI database: NM_170596) that includes exons 1–3. The ≈150-kDa signal seems independent of the full length protein and may represent a highly stable complex (27) involving shorter Disc1 variant(s) or a nonspecific signal. C57, C57BL/6J; 129, 129S6/SvEv. (c) Predicted protein domains of the mDISC1 protein from C57BL/6J and 129S6/SvEv strains. Black boxes indicate predicted coiled-coil domains identified by COILS search (
www.ch.embnet.org/software/COILS-form.html
). Upper horizontal lines indicate the predicted leucine zipper domain. Open bar indicates the approximate position of the peptide antigen. BP, approximate position of the truncation in the human orthologue induced by the breakpoint in the Scottish family.
Fig. 3.
mDisc1 deficiency results in impaired working memory performance. (a) Gross brain morphology in _mDisc1_-deficient mice. Shown are cresyl violet-stained coronal sections through the brain of adult _mDisc1_-deficient mice (Hom) and their WT littermates (Wt). (Top) Prefrontal cortex at ≈−1.70 mm Bregma. (Middle) Cortex at ≈−2.18 mm Bregma. (Bottom) Hippocampus. (b) Working memory performance in a delayed non-match to place task. (Left) Mean percentage of correct responses (± SEM) for WT (Wt), heterozygous (Het), and homozygous (Hom) mice. Note that vertical axis starts at 50% correct responses, which represents baseline response accuracy expected by chance. (Right) Mean time (± SEM) to complete the working memory task measured in seconds. There were no differences between genotypes either in the number of days to reach criterion or in the level of performance in the 3 days before testing. (c) Spontaneous locomotion. (Left) Total distance traveled (± SEM) as a function of time in 3-min bins. (Right) Distance traveled (± SEM) over a 30-min period in the margins and center of a novel open field environment. (d) Prepulse inhibition. Percentage inhibition (± SEM) at different levels of prepulse intensities. There were no observed differences in background levels or startle responses.
Similar articles
- Phenotypic characterization of C57BL/6J mice carrying the Disc1 gene from the 129S6/SvEv strain.
Juan LW, Liao CC, Lai WS, Chang CY, Pei JC, Wong WR, Liu CM, Hwu HG, Lee LJ. Juan LW, et al. Brain Struct Funct. 2014 Jul;219(4):1417-31. doi: 10.1007/s00429-013-0577-8. Epub 2013 May 21. Brain Struct Funct. 2014. PMID: 23689501 - Frontal cortical synaptic communication is abnormal in Disc1 genetic mouse models of schizophrenia.
Holley SM, Wang EA, Cepeda C, Jentsch JD, Ross CA, Pletnikov MV, Levine MS. Holley SM, et al. Schizophr Res. 2013 May;146(1-3):264-72. doi: 10.1016/j.schres.2013.02.007. Epub 2013 Mar 6. Schizophr Res. 2013. PMID: 23481583 Free PMC article. - Comprehensive behavioral analysis of ENU-induced Disc1-Q31L and -L100P mutant mice.
Shoji H, Toyama K, Takamiya Y, Wakana S, Gondo Y, Miyakawa T. Shoji H, et al. BMC Res Notes. 2012 Feb 20;5:108. doi: 10.1186/1756-0500-5-108. BMC Res Notes. 2012. PMID: 22348257 Free PMC article. - Deletion polymorphism of Disc1 is common to all 129 mouse substrains: implications for gene-targeting studies of brain function.
Clapcote SJ, Roder JC. Clapcote SJ, et al. Genetics. 2006 Aug;173(4):2407-10. doi: 10.1534/genetics.106.060749. Epub 2006 Jun 4. Genetics. 2006. PMID: 16751659 Free PMC article. Review. - DISC1 in schizophrenia: genetic mouse models and human genomic imaging.
Johnstone M, Thomson PA, Hall J, McIntosh AM, Lawrie SM, Porteous DJ. Johnstone M, et al. Schizophr Bull. 2011 Jan;37(1):14-20. doi: 10.1093/schbul/sbq135. Epub 2010 Dec 13. Schizophr Bull. 2011. PMID: 21149852 Free PMC article. Review.
Cited by
- Hippocampal place cell and inhibitory neuron activity in disrupted-in-schizophrenia-1 mutant mice: implications for working memory deficits.
Mesbah-Oskui L, Georgiou J, Roder JC. Mesbah-Oskui L, et al. NPJ Schizophr. 2015 Apr 1;1:15011. doi: 10.1038/npjschz.2015.11. eCollection 2015. NPJ Schizophr. 2015. PMID: 27336029 Free PMC article. - Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1.
Hayashi-Takagi A, Takaki M, Graziane N, Seshadri S, Murdoch H, Dunlop AJ, Makino Y, Seshadri AJ, Ishizuka K, Srivastava DP, Xie Z, Baraban JM, Houslay MD, Tomoda T, Brandon NJ, Kamiya A, Yan Z, Penzes P, Sawa A. Hayashi-Takagi A, et al. Nat Neurosci. 2010 Mar;13(3):327-32. doi: 10.1038/nn.2487. Epub 2010 Feb 7. Nat Neurosci. 2010. PMID: 20139976 Free PMC article. - Animal models of gene-environment interactions in schizophrenia.
Ayhan Y, Sawa A, Ross CA, Pletnikov MV. Ayhan Y, et al. Behav Brain Res. 2009 Dec 7;204(2):274-81. doi: 10.1016/j.bbr.2009.04.010. Epub 2009 Apr 18. Behav Brain Res. 2009. PMID: 19379776 Free PMC article. - The effects of arousal on apical amplification and conscious state.
Phillips WA, Larkum ME, Harley CW, Silverstein SM. Phillips WA, et al. Neurosci Conscious. 2016 Sep 11;2016(1):niw015. doi: 10.1093/nc/niw015. eCollection 2016. Neurosci Conscious. 2016. PMID: 29877512 Free PMC article. Review. - Impaired long-term memory retention and working memory in sdy mutant mice with a deletion in Dtnbp1, a susceptibility gene for schizophrenia.
Takao K, Toyama K, Nakanishi K, Hattori S, Takamura H, Takeda M, Miyakawa T, Hashimoto R. Takao K, et al. Mol Brain. 2008 Oct 22;1:11. doi: 10.1186/1756-6606-1-11. Mol Brain. 2008. PMID: 18945333 Free PMC article.
References
- Gogos J. A., Gerber D. J. Trends Pharmacol. Sci. 2006 in press. - PubMed
- Millar J. K., Wilson-Annan J. C., Anderson S., Christie S., Taylor M. S., Semple C. A., Devon R. S., Clair D. M., Muir W. J., Blackwood D. H., et al. Hum. Mol. Genet. 2000;9:1415–1423. - PubMed
- Ekelund J., Hennah W., Hiekkalinna T., Parker A., Meyer J., Lonnqvist J., Peltonen L. Mol. Psychiatry. 2004;9:1037–1041. - PubMed
- Hennah W., Tuulio-Henriksson A., Paunio T., Ekelund J., Varilo T., Partonen T., Cannon T. D., Lonnqvist J., Peltonen L. Mol. Psychiatry. 2005;10:1097–1103. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases