RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules - PubMed (original) (raw)
RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules
Wei-Hong Yang et al. RNA. 2006 Apr.
Abstract
The mRNA processing body (P-body) is a cellular structure that has an important role in mRNA degradation. P-bodies have also been implicated in RNAi-mediated post-transcriptional gene silencing. The objective of this study was to identify and characterize novel components of the mammalian P-body. Approximately 5% of patients with the autoimmune disease primary biliary cirrhosis have antibodies directed against this structure. Serum from one of these patients was used to identify a cDNA encoding RAP55, a 463-amino acid protein. RAP55 colocalized with previously identified P-body components DCP1a and Ge-1. RAP55 contains an N-terminal Sm-like domain and two C-terminal RGG-rich domains separated by an FDF motif. The two RGG domains and the FDF domain were necessary and sufficient to target the protein to P-bodies. A fragment of RAP55 consisting of the FDF and the second RGG domains did not localize to P-bodies, but was able to displace other P-body components from this structure. After cells were subjected to arsenite-induced stress, RAP55 was detected in TIA-containing stress granules. The second RGG domain was necessary and sufficient for stress granule localization. siRNA-mediated knock-down of RAP55 resulted in loss of P-bodies, suggesting that RAP55 acts prior to the 5'-decapping step in mRNA degradation. The results of this study show that RAP55 is a component of P-bodies in cells at rest and localizes in stress granules in arsenite-treated cells. RAP55 may serve to shuttle mRNAs between P-bodies and stress granules.
Figures
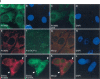
FIGURE 1.
Antibodies in the serum of a primary biliary cirrhosis patient react with P-bodies and stress granules. (Panel I) (A,E) Patient 0080’s serum contained antibodies that reacted with 5–20 dots in the cytoplasm of Hep-2 cells as determined by indirect immunofluorescence. The patient’s serum also contained antibodies directed against E2-pyruvate dehydrogenase complex (E2-PDC), which produced granular, filamentous (mitochondrial) cytoplasmic staining. (C) Treatment of Hep-2 cells with cyclohexamide resulted in disappearance of cytoplasmic dots with no effect on the staining of mitochondria. (E) Patient antibodies colocalized with (F) anti-DCP1a antibodies. To examine the effect of stress on the cellular location of the P-body antigen, cells were treated with arsenite and stained with (I) human serum and (J) anti-TIA antiserum. Human serum 0080 reacted with one or more proteins that colocalized with TIA in stress granules. Overlap of red and green staining is shown in yellow in G and K. DAPI staining in B, D, H, and L indicates the location of cell nuclei. (Panel II) Immunoblotting was used to characterize the putative P-body/stress granule autoantigen. (A) Antibodies in patient 0080’s serum reacted with 70-kDa (E2-PDC) and 60-kDa proteins in an extract prepared from Hep-2 cells. In addition, the serum reacted weakly with a 50-kDa band, which may be a breakdown product of one of the larger proteins (lane 1). Serum from patient 0012 reacted with the 60-kDa protein, but not with E2 PDC. Serum from a third patient (Ge) reacted with a 160-kDa protein, previously shown to be Ge-1, as well as E2 PDC. In addition, Ge serum produced weak bands at 60 kDa and 50 kDa. Serum from patient 0050 reacted with Ge-1 and an unknown 110-kDa protein (a black arrow indicates the location of this protein). (B) To determine the cellular location of the 110-kDa protein, serum 0050 was used to probe an immunoblot prepared using extracts from cytoplasmic (Cyt) or nuclear (Nuc) fractions of Hep-2 cells. Ge-1 was detected in the cytoplasmic fraction, and the unidentified 110-kDa autoantigen was present in the nuclear fraction.
FIGURE 1.
Antibodies in the serum of a primary biliary cirrhosis patient react with P-bodies and stress granules. (Panel I) (A,E) Patient 0080’s serum contained antibodies that reacted with 5–20 dots in the cytoplasm of Hep-2 cells as determined by indirect immunofluorescence. The patient’s serum also contained antibodies directed against E2-pyruvate dehydrogenase complex (E2-PDC), which produced granular, filamentous (mitochondrial) cytoplasmic staining. (C) Treatment of Hep-2 cells with cyclohexamide resulted in disappearance of cytoplasmic dots with no effect on the staining of mitochondria. (E) Patient antibodies colocalized with (F) anti-DCP1a antibodies. To examine the effect of stress on the cellular location of the P-body antigen, cells were treated with arsenite and stained with (I) human serum and (J) anti-TIA antiserum. Human serum 0080 reacted with one or more proteins that colocalized with TIA in stress granules. Overlap of red and green staining is shown in yellow in G and K. DAPI staining in B, D, H, and L indicates the location of cell nuclei. (Panel II) Immunoblotting was used to characterize the putative P-body/stress granule autoantigen. (A) Antibodies in patient 0080’s serum reacted with 70-kDa (E2-PDC) and 60-kDa proteins in an extract prepared from Hep-2 cells. In addition, the serum reacted weakly with a 50-kDa band, which may be a breakdown product of one of the larger proteins (lane 1). Serum from patient 0012 reacted with the 60-kDa protein, but not with E2 PDC. Serum from a third patient (Ge) reacted with a 160-kDa protein, previously shown to be Ge-1, as well as E2 PDC. In addition, Ge serum produced weak bands at 60 kDa and 50 kDa. Serum from patient 0050 reacted with Ge-1 and an unknown 110-kDa protein (a black arrow indicates the location of this protein). (B) To determine the cellular location of the 110-kDa protein, serum 0050 was used to probe an immunoblot prepared using extracts from cytoplasmic (Cyt) or nuclear (Nuc) fractions of Hep-2 cells. Ge-1 was detected in the cytoplasmic fraction, and the unidentified 110-kDa autoantigen was present in the nuclear fraction.
FIGURE 2.
Structure of RAP55 and identification of the portions of RAP55 that mediate localization to P-bodies and stress granules. (Panel I) The predicted amino acid sequence of RAP55 contains an N-terminal LSm domain and two C-terminal RGG-rich domains. Between the two RGG domains is an FDF motif. The portions of RAP55 that were capable of localizing green fluorescent protein (GFP) to P-bodies (PB) or stress granules (SG) are indicated by +. Representative indirect immunofluorescence results are shown in panel II. (Panel II) Two-color immunofluorescence was used to identify the portions of RAP55 that were necessary and sufficient for localization to P-bodies. (A) Full-length GFP-RAP55 localized to cytoplasmic dots and (B) colocalized with cotransfected DCP1a. (C) Rabbit anti-RAP55 antibodies colocalized with Ge-1-containing P-bodies as determined (D) using human serum 0050, which reacts with Ge-1, but not RAP55. A C-terminal portion of RAP55 (amino acids 265–463) (E) localized to P-bodies and (F) colocalized with Ge-1. White arrows indicate examples of overlapping cytoplasmic foci. (G) RAP55(256–403), which lacks the RGG2 domain, was present diffusely throughout the cytoplasm and did not localize to P-bodies. (H) RAP55(256–403) did not alter the P-body location of Ge-1. (I) RAP55(289–463) also did not localize to P-bodies, but this portion of RAP55 (J) displaced Ge-1 from P-bodies. Similarly, (K) RAP55(289–463) displaced DCP1a from (L) P-bodies in transfected cells. Note that DCP1a-containing P-bodies were detected in nontransfected cells (indicated by white arrows in K and L). Successful production of the GFP-RAP55 fragment fusion proteins used in these studies was confirmed by immunoblot using anti-GFP antiserum (data not shown).
FIGURE 2.
Structure of RAP55 and identification of the portions of RAP55 that mediate localization to P-bodies and stress granules. (Panel I) The predicted amino acid sequence of RAP55 contains an N-terminal LSm domain and two C-terminal RGG-rich domains. Between the two RGG domains is an FDF motif. The portions of RAP55 that were capable of localizing green fluorescent protein (GFP) to P-bodies (PB) or stress granules (SG) are indicated by +. Representative indirect immunofluorescence results are shown in panel II. (Panel II) Two-color immunofluorescence was used to identify the portions of RAP55 that were necessary and sufficient for localization to P-bodies. (A) Full-length GFP-RAP55 localized to cytoplasmic dots and (B) colocalized with cotransfected DCP1a. (C) Rabbit anti-RAP55 antibodies colocalized with Ge-1-containing P-bodies as determined (D) using human serum 0050, which reacts with Ge-1, but not RAP55. A C-terminal portion of RAP55 (amino acids 265–463) (E) localized to P-bodies and (F) colocalized with Ge-1. White arrows indicate examples of overlapping cytoplasmic foci. (G) RAP55(256–403), which lacks the RGG2 domain, was present diffusely throughout the cytoplasm and did not localize to P-bodies. (H) RAP55(256–403) did not alter the P-body location of Ge-1. (I) RAP55(289–463) also did not localize to P-bodies, but this portion of RAP55 (J) displaced Ge-1 from P-bodies. Similarly, (K) RAP55(289–463) displaced DCP1a from (L) P-bodies in transfected cells. Note that DCP1a-containing P-bodies were detected in nontransfected cells (indicated by white arrows in K and L). Successful production of the GFP-RAP55 fragment fusion proteins used in these studies was confirmed by immunoblot using anti-GFP antiserum (data not shown).
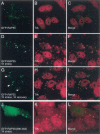
FIGURE 3.
The effect of arsenite-induced oxidative stress on the cellular location of RAP55. (A) In resting Hep-2 cells, RAP55-containing P-bodies were distributed throughout the cytoplasm, and (B) TIA was detected diffusely throughout the nucleus, but excluded from nucleoli. After 60 min of exposure to sodium arsenite, both (D) RAP55 and (E) TIA were detected in cytoplasmic granules. (H) After 1 h exposure to arsenite and 1 h recovery in culture medium, TIA-containing stress granules persisted in the cell cytoplasm. RAP55 was detected both within stress granules and in adjacent P-bodies (indicated by white arrows in G). To identify the portion of RAP55 that mediated localization to stress granules, GFP-RAP55 fragment fusion proteins were expressed in Hep-2 cells, which were then subjected to arsenite-induced stress for 1 h. (J) RAP55 amino acids 396–463 were necessary and sufficient to direct GFP to (K) TIA-containing stress granules. Overlap of green and red staining appears yellow in C, F, I, and L.
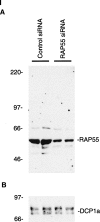
FIGURE 4.
The effect of RAP55 knock-down on P-bodies. (Panel I) Immunoblotting was used to determine the effect of siRNAs on the levels of RAP55 and DCP1a in Hep-2 cells. Compared with control siRNA, RAP55 siRNA decreased the level of (A) RAP55 but not (B) DCP1a in Hep-2 cells. As indicated, siRNA knock-down was performed in duplicate. RAP55 and DCP1a were detected using patient serum 0080 and rabbit anti-DCP1a antiserum, respectively. The human serum reacted weakly with E2 PDC (70 kDa). (Panel II) RAP55 siRNA decreased the level of RAP55, DCP1a, and Ge-1 in P-bodies. After transfection of RAP55 siRNA into Hep-2 cells, neither (A) RAP55 nor (B) DCP1a was detected in P-bodies. (D,E) Control siRNA did not alter the P-body location of either protein. (G,H) Transfection of RAP55 siRNA into Hep-2 cells also displaced Ge-1 from P-bodies. (J,K) Control siRNA had no effect on the P-body localization of Ge-1. Patient 0012 serum or rabbit anti-RAP55 antiserum was used to detect RAP55, as indicated. Patient 0050 serum was used to detect Ge-1. DAPI staining indicates the location of nuclei in C, F, I, and L.
FIGURE 4.
The effect of RAP55 knock-down on P-bodies. (Panel I) Immunoblotting was used to determine the effect of siRNAs on the levels of RAP55 and DCP1a in Hep-2 cells. Compared with control siRNA, RAP55 siRNA decreased the level of (A) RAP55 but not (B) DCP1a in Hep-2 cells. As indicated, siRNA knock-down was performed in duplicate. RAP55 and DCP1a were detected using patient serum 0080 and rabbit anti-DCP1a antiserum, respectively. The human serum reacted weakly with E2 PDC (70 kDa). (Panel II) RAP55 siRNA decreased the level of RAP55, DCP1a, and Ge-1 in P-bodies. After transfection of RAP55 siRNA into Hep-2 cells, neither (A) RAP55 nor (B) DCP1a was detected in P-bodies. (D,E) Control siRNA did not alter the P-body location of either protein. (G,H) Transfection of RAP55 siRNA into Hep-2 cells also displaced Ge-1 from P-bodies. (J,K) Control siRNA had no effect on the P-body localization of Ge-1. Patient 0012 serum or rabbit anti-RAP55 antiserum was used to detect RAP55, as indicated. Patient 0050 serum was used to detect Ge-1. DAPI staining indicates the location of nuclei in C, F, I, and L.
Similar articles
- Probing the mRNA processing body using protein macroarrays and "autoantigenomics".
Yang WH, Bloch DB. Yang WH, et al. RNA. 2007 May;13(5):704-12. doi: 10.1261/rna.411907. Epub 2007 Mar 5. RNA. 2007. PMID: 17339575 Free PMC article. - PRMT1 is required for RAP55 to localize to processing bodies.
Matsumoto K, Nakayama H, Yoshimura M, Masuda A, Dohmae N, Matsumoto S, Tsujimoto M. Matsumoto K, et al. RNA Biol. 2012 May;9(5):610-23. doi: 10.4161/rna.19527. Epub 2012 May 1. RNA Biol. 2012. PMID: 22614839 - The NS1 protein of influenza A virus interacts with cellular processing bodies and stress granules through RNA-associated protein 55 (RAP55) during virus infection.
Mok BW, Song W, Wang P, Tai H, Chen Y, Zheng M, Wen X, Lau SY, Wu WL, Matsumoto K, Yuen KY, Chen H. Mok BW, et al. J Virol. 2012 Dec;86(23):12695-707. doi: 10.1128/JVI.00647-12. Epub 2012 Sep 12. J Virol. 2012. PMID: 22973032 Free PMC article. - RAP55: insights into an evolutionarily conserved protein family.
Marnef A, Sommerville J, Ladomery MR. Marnef A, et al. Int J Biochem Cell Biol. 2009 May;41(5):977-81. doi: 10.1016/j.biocel.2008.06.015. Epub 2008 Aug 3. Int J Biochem Cell Biol. 2009. PMID: 18723115 Review. - Relationship of other cytoplasmic ribonucleoprotein bodies (cRNPB) to GW/P bodies.
Moser JJ, Fritzler MJ. Moser JJ, et al. Adv Exp Med Biol. 2013;768:213-42. doi: 10.1007/978-1-4614-5107-5_13. Adv Exp Med Biol. 2013. PMID: 23224973 Review.
Cited by
- Eukaryotic mRNA decapping factors: molecular mechanisms and activity.
He F, Jacobson A. He F, et al. FEBS J. 2023 Nov;290(21):5057-5085. doi: 10.1111/febs.16626. Epub 2022 Sep 30. FEBS J. 2023. PMID: 36098474 Free PMC article. Review. - Who Regulates Whom? An Overview of RNA Granules and Viral Infections.
Poblete-Durán N, Prades-Pérez Y, Vera-Otarola J, Soto-Rifo R, Valiente-Echeverría F. Poblete-Durán N, et al. Viruses. 2016 Jun 28;8(7):180. doi: 10.3390/v8070180. Viruses. 2016. PMID: 27367717 Free PMC article. Review. - Hepatitis C virus infection alters P-body composition but is independent of P-body granules.
Pérez-Vilaró G, Scheller N, Saludes V, Díez J. Pérez-Vilaró G, et al. J Virol. 2012 Aug;86(16):8740-9. doi: 10.1128/JVI.07167-11. Epub 2012 Jun 6. J Virol. 2012. PMID: 22674998 Free PMC article. - Spinal muscular atrophy and a model for survival of motor neuron protein function in axonal ribonucleoprotein complexes.
Rossoll W, Bassell GJ. Rossoll W, et al. Results Probl Cell Differ. 2009;48:289-326. doi: 10.1007/400_2009_4. Results Probl Cell Differ. 2009. PMID: 19343312 Free PMC article. - Crystal structure of human Edc3 and its functional implications.
Ling SH, Decker CJ, Walsh MA, She M, Parker R, Song H. Ling SH, et al. Mol Cell Biol. 2008 Oct;28(19):5965-76. doi: 10.1128/MCB.00761-08. Epub 2008 Aug 4. Mol Cell Biol. 2008. PMID: 18678652 Free PMC article.
References
- Bloch, D.B., Rabkina, D., Quertermous, T., and Bloch, K.D. 1994. The immunoreactive region in a novel autoantigen contains a nuclear localization sequence. Clin. Immunol. Immunopathol. 72: 380–389. - PubMed
- Bloch, D.B., Yu, J.H., Yang, W.H., Graeme-Cook, F., Lindor, K.D., Viswanathan, A., Bloch, K.D., and Nakajima, A. 2005. The cytoplasmic dot staining pattern is detected in a subgroup of patients with primary biliary cirrhosis. J. Rheumatol. 32: 477–483. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL074352/HL/NHLBI NIH HHS/United States
- HL070896/HL/NHLBI NIH HHS/United States
- R01 HL070896/HL/NHLBI NIH HHS/United States
- DK051179/DK/NIDDK NIH HHS/United States
- HL074352/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases