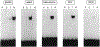Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors - PubMed (original) (raw)
Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors
K Umesono et al. Cell. 1991.
Abstract
We report here the identification of thyroid hormone response elements (TREs) that consist of a direct repeat, not a palindrome, of the half-sites. Unlike palindromic TREs, direct repeat TREs do not confer a retinoic acid response. The tandem TRE can be converted into a retinoic acid response element by increasing the spacing between the half-sites by 1 nucleotide, and the resulting retinoic acid response element is no longer a TRE. Decreasing the half-site spacing by 1 nucleotide converts the TRE to a vitamin D3 response element, while eliminating response to T3. These results correlate well with DNA-binding affinities of the thyroid hormone, retinoic acid, and vitamin D3 receptors. This study points to the general importance of tandem repeat hormone response elements and suggests a simple physiologic code exists in which half-site spacing plays a critical role in achieving selective hormonal response.
Figures
Figure 1.. TR-Specific Transactivation of αTRE-CAT
The reporter plasmid pαTRE.Al0-CAT encodes a portion of the rat cardiac MHC α subunit gene, which confers T3 responsiveness onto a heterologous promoter to drive expression of a chloramphenicol acetyltransferase (CAT) gene (Izumo and Mahdavi. 1988). Together with an internal control plasmid (pRAS-βGAL), the reporter plasmid was cotransfected with an expression plasmid coding for the firefly luciferase (−), the human RARα (hRARα), -β (hRARβ). or -γ (hRAR γ), or the human TRβ (hTRβ) into CV-1 cells. Subsequently, cognate hormones were added to the culture media at 1 μM RA, 100 nM T3, or solvent ethanol (−). Thirty-six hours after adding the ligands, cellular extracts were prepared and assayed for the CAT enzyme activity after monitoring the β-galactosidase activity to correct variation of the transfection efficiency.
Figure 2.. Structure and Responsiveness of Target HREs for T3 and RA
Target HREs for the TR and RAR were synthesized as double-stranded oligonucleotides with an overhanging tetranucleotide (5’-agct-3’) at both ends. A single copy of these oligonucleotides was cloned at the unique Hindlll site present in a basal promoter CAT construct, ΔSV-CAT (see Experimental Procedures). Capitalized portions in the nucleotide sequences correspond to those found in the natural promoters except TREp and rGH21, which are synthetic. Bold letters and arrows indicate the AGGTCA motif and “X” denotes a nucleotide substitution from this motif. Numbers between the arrows are the size of the spacer and those in columns represent fold inductions of the CAT enzyme activity stimulated by the hormones in either the TR8groducing (± T3 at 100 nM) or RARa-producing (± RA at 1 μM) CV-1 cells. Inductions observed on the basal construct ΔSV-CAT by T3 and RA are 0.8 and 1 4-fold, respectively. (a) TREp is an optimized palindromic rGH TRE (Glass et al., 1988). which stands also as an efficient RARE (Umesono et al., 1988). MHC-L encodes a TRE localized at positions between −154 and −122 from a transcription start site in the MHC gene promoter (shown as an antisense; Glass et al., 1989). MHC-S and -D contain deletion(s) (indicated by brackets) from the MHC-L. (b) MHC-N encodes a core sequence of the wild-type MHC TRE, whereas Ml through M3 contain specific nucleotide substitutions (shaded letters and “X” in the arrow if they are located in the AGGTCA motif). (c) Malic enzyme TRE corresponds to −281 to −261 from a transcription start site (Petty et al., 1990). A TRE found in the MLV LTR was taken from Sap et al. (1989) and rGH21 is a mutant of the rGH gene TRE (Brent et al., 1989a). βRARE corresponds to 8RE2 reported in Sucov et al. (1990).
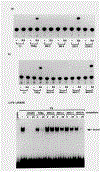
Figure 3.. In Vitro DNA Binding of the TRβ Protein to MHC TRE Mutants
As a radiolabeled probe for the gel retardation DNA-binding assay, the MHC-N double-stranded oligonucleotides were labeled by a filling-in reaction with Klenow enzyme in the presence of [α−32P]dCTP. TRβ protein was overexpressed in COS cells by transfecting 20 μg of pCMX-TRβ plasmid. As a control, cell extracts were also prepared from mock-transfected (20 μg of pCMX-LUC) COS cells. Protein (5 μg) of the mock- (control) or TRβ-transfected cell extracts was incubated with the MHC-N probe, and the protein-DNA complex was resolved electrophoretically according to Damm et al. (1989). With the TRβ extracts, 50-fold excess amounts of cold oligonucleotides as indicated were added as a competitor for the TRβ binding to the MHC-N.
Figure 4.. Mutually Exclusive HRE Binding by the TRβ and RARα Proteins
The32P-labeled oligonucleotides at the top (listed in Figure 2, αMHC corresponds to MHC-N oligonucleotides) were used as a probe for gel retardation DNA-binding assays with 5 μg of protein of either control (C), TRβ (T), or RARα (R) COS cell extracts. These extracts were prepared aftertransfecting COScells with 20 μg of pCMX-LUC, pCMX-hTRβ, or pCMX-hRARα expression plasmids, respectively.
Figure 5.. Conversion of the MHC TRE into an RARE
(a) Four different oligonucleotides were synthesized encoding the wild-type MHC TRE (MHC-N), an insertion mutant in the spacer region (MHC+l), a half-site mutant (MHC-T), and a double mutant carrying both the MHC+l and MHC-T mutations (MHC-R). A single copy of these oligonucleotides was placed in the basal reporter construct ΔSV-CAT (−), giving rise to a set of CAT reporter plasmids together with one encoding TREp (see Figure 2). (b) Transactivation assays by T3 and RA. CV-1 cells were transfected with one of these reporters along with an expression plasmid coding for the TRβ or RARα. After addition of the hormones (RA, 1 μM; T3, 100 nM), the cell extracts were assayed for the CAT activity by measuringthe control β-galactosidase activity produced by cotransfected pRAS-βGAL. Numbers in the columns indicate a fold induction of the CAT activity by the hormone.
Figure 6.. In Vitro DNA Binding of the TRβ and RARα to MHC TRE/RARE Mutants
32P-labeled MHC-N (a) or βRARE (b) oligonucleotides were used as a probe for gel retardation DNA-binding assays with 5 μg of protein of either control, TRβ. or RARα cell extracts as in Figure 4. A minus sign indicates no competitor was included during the reaction. Otherwise, indicated oligonucleotides (βRARE, TREp, MHC-N, MHC+l, MHC-T, MHC-R) with either 5- or 25-fold molar excess to the probe were added to the binding reaction.
Figure 7.. Selecttve Transactivation of Synthetic Direct Repeat HREs by VD3, T3. and RA
(a) Nucleotide sequences of the oligonucleo-tides encoding synthetic direct repeat HREs. DR-3, −4, and −5 code for a perfect tandem repeat of AGGTCA hextamers (indicated by arrows) separated by 3, 4, and 5 nucleotides, respectively. GDR-3, −4, and −5 are identical to the DR oligonucleotides except that the half-site sequence was changed to AG
AA
CA, a GRE half-site. (b-d) A single copy of DR or GDR oligonucleo-tides was cloned at the unique Hindlll site present in the basal promoter CAT construct ΔSVCAT, giving rise to DR-3-CAT, DR-CCAT, DR-SCAT. GDR-3CAT. GDR-4CAT, and GDR-5CAT reporters. One mtcrogram of the indicated reporters (a minus sign corresponds to ASV-CAT) was cotransfected along with 0.5 μg of an expression plasmid for VDR (b), TRβ (c). or RARα (d) to CV-1 cells. After 36 hr of incubation with the cognate ligands (VD3, and T3; 100 nM; RA, 1 μM), the cells were harvested for the CAT assay after normalization with β-galactosidase activity produced from the cotransfected control reporter pRAS-βGAL. The CAT activity obtained through ΔSV-CAT in the absence of the ligand was taken as 1 for each of the receotors.
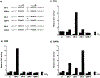
Figure 8.. RAeDependent Transactivation in F9 Teratocarcinoma Cells
(a and b) Indicated reporter CAT plasmids (2 μg) were transfected into F9 teratocarcinoma cells with the control pRA-βGAL plasmid. After 24 hr of incubation with RA at 1 μM, the cells were harvested and CAT activities were analyzed. (c) F9 cells were kept undifferentiated in the media and whole-cell extracts were prepared with 0.4 M KCI. We have confirmed that the F9 cells express RARα and RARγ, but not RARβ mRNAs (A. Kakizuka and R. M. E., unpublished data). Ten micrograms of the F9 proteins was incubated with 40 fmol of 32P-labeled f3RARE oligonucleotide for protein-DNA binding assays. When included, either a 5- or 25-fold molar excess of unlabeled competitor oligonucleotides (βRARE, TREp, MHC-N, MHC+i, MHC-T, or MHC-R) to the probe was added to the reaction mixture.
Figure 9.. Specific DNA Binding of VDR, TRβ, and RARα Proteins to the Direct Repeat HREs
[α−32P]dCTP-labeled DR-3 (a), DR-4 (b), or DR-5 (c) oligonucleotides were used as a probe for gel retardation DNA-binding assays with 5 μg of protein of either control, VDR-, TRβ-, or RARα-transfected COS cell extracts as in Figure 4. A minus sign indicates no competitor was included during the binding reaction. Otherwise, indicatedoligonucle tides with either 5- or 25-fold molar excess to the probe were added to the reaction mixtures as a competitor.
Figure 10.. Sequence Comparison of the Direct Repeat HREs
(a) rOST, the rat osteocalcin gene promoter (−455 to −441 from a transcription start site; Demay et al., 1990); hOST, the human osteocalcin gene promoter (−499 to −485; Ozono et al., 1991); mSPP-1, the mouse Sppllosteopontin promoter (−757 to −743; Noda et al., 1990). We confirmed that this spp-l VDREconfers vitamin D3 response io the basal reporter ΔSV-CAT, but a similar motif found in the 3’ end of the mouse laminin Bl RARE (mLamB1, −448 to −433 shown as an antisense after Vasios et al., 1989) does not function as a VDRE (data not shown). (b) rMHC, the rat cardiac MHC α gene promoter (−149 to −134; Izumo and Mahdavi, 1988; Glass et al., 1989); hMHC, the human MHC promoter (−158 to −143; Flink and Morkin, 1990); rME, the rat malic enzyme gene promoter (−276 to −261; Petty et al., 1990); MLV, positions between 334 and 350 shown as an antisense (Sap et al., 1989); rS14, the rat S14 gene TRE (personal communication from How ard C. Towle). The oligonucleotides encoding DR-4M (A toT change in the 5’ end of each of the half-sites in DR-4) or the mouse skeletat actin gene promoter upstream sequence (mACTIN, −597 to −582 from a transcription start site; Hu et al., 1986) failed to confer T3 response to the basal reporter (data not shown). For rGH21 and βRARE-1, see text. (c) mRARβ, the mouse RAR type β gene promoter (Sucov et al., 1990); hRARβ, the human RAR type β gene promoter (−52 to −37; de The et al., 1990); mCP-H, the mouse complement factor H gene promoter (−143 to −127; Muiioz-Cdnoves et al., 1990); hADH3, the human alcohol dehydrogenase gene promoter (−300 to −284 shown as an antisense; Duester et al., 1991). Oligonucleotides coding for a motif found in 2.4 kb upstream of the human muscle creatine kinase gene (hMCK; Trask et al., 1988) did not confer RA response (data not shown).
Similar articles
- RXR beta: a coregulator that enhances binding of retinoic acid, thyroid hormone, and vitamin D receptors to their cognate response elements.
Yu VC, Delsert C, Andersen B, Holloway JM, Devary OV, Näär AM, Kim SY, Boutin JM, Glass CK, Rosenfeld MG. Yu VC, et al. Cell. 1991 Dec 20;67(6):1251-66. doi: 10.1016/0092-8674(91)90301-e. Cell. 1991. PMID: 1662118 - The interplay of half-site sequence and spacing on the activity of direct repeat thyroid hormone response elements.
Katz RW, Subauste JS, Koenig RJ. Katz RW, et al. J Biol Chem. 1995 Mar 10;270(10):5238-42. doi: 10.1074/jbc.270.10.5238. J Biol Chem. 1995. PMID: 7890633 - The erb-A family receptors for thyroid hormones, steroids, vitamin D and retinoic acid: characteristics and modulation.
Nunez EA. Nunez EA. Curr Opin Cell Biol. 1989 Apr;1(2):177-85. doi: 10.1016/0955-0674(89)90083-5. Curr Opin Cell Biol. 1989. PMID: 2561067 Review. No abstract available. - How do thyroid hormone receptors bind to structurally diverse response elements?
Desvergne B. Desvergne B. Mol Cell Endocrinol. 1994 Apr;100(1-2):125-31. doi: 10.1016/0303-7207(94)90291-7. Mol Cell Endocrinol. 1994. PMID: 8056146 Review.
Cited by
- Vitamin D and Sarcopenia in the Senior People: A Review of Mechanisms and Comprehensive Prevention and Treatment Strategies.
Zhang F, Li W. Zhang F, et al. Ther Clin Risk Manag. 2024 Sep 5;20:577-595. doi: 10.2147/TCRM.S471191. eCollection 2024. Ther Clin Risk Manag. 2024. PMID: 39253031 Free PMC article. Review. - Thyroid hormone-regulated chromatin landscape and transcriptional sensitivity of the pituitary gland.
Cho YW, Fu Y, Huang CJ, Wu X, Ng L, Kelley KA, Vella KR, Berg AH, Hollenberg AN, Liu H, Forrest D. Cho YW, et al. Commun Biol. 2023 Dec 11;6(1):1253. doi: 10.1038/s42003-023-05546-y. Commun Biol. 2023. PMID: 38081939 Free PMC article. - Nuclear retinoid receptors and pregnancy: placental transfer, functions, and pharmacological aspects.
Comptour A, Rouzaire M, Belville C, Bouvier D, Gallot D, Blanchon L, Sapin V. Comptour A, et al. Cell Mol Life Sci. 2016 Oct;73(20):3823-37. doi: 10.1007/s00018-016-2332-9. Epub 2016 Aug 9. Cell Mol Life Sci. 2016. PMID: 27502420 Free PMC article. Review. - Design of Large-Scale Reporter Construct Arrays for Dynamic, Live Cell Systems Biology.
Decker JT, Hall MS, Peñalver-Bernabé B, Blaisdell RB, Liebman LN, Jeruss JS, Shea LD. Decker JT, et al. ACS Synth Biol. 2018 Sep 21;7(9):2063-2073. doi: 10.1021/acssynbio.8b00236. Epub 2018 Sep 10. ACS Synth Biol. 2018. PMID: 30189139 Free PMC article. - Retinoic acid receptor-mediated induction of ABCA1 in macrophages.
Costet P, Lalanne F, Gerbod-Giannone MC, Molina JR, Fu X, Lund EG, Gudas LJ, Tall AR. Costet P, et al. Mol Cell Biol. 2003 Nov;23(21):7756-66. doi: 10.1128/MCB.23.21.7756-7766.2003. Mol Cell Biol. 2003. PMID: 14560020 Free PMC article.
References
- Baniahmad A, Steiner C, Kohne AC, and Renkawitz R (1990). Modular structure of a chicken lysozyme silencer: involvement of an unusual thyroid hormone receptor binding site. Cell 67, 505–514. - PubMed
- Beato M (1989). Gene regulation by steroid hormones. Cell 56335–344. - PubMed
- Benbrook D, Lernhardt E, and Pfahl M (1988). A new retinoic acid receptor identified from a hepatocellular carcinoma. Nature 333,669–672. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources