Euchromatin and pericentromeric heterochromatin: comparative composition in the tomato genome - PubMed (original) (raw)
Comparative Study
Euchromatin and pericentromeric heterochromatin: comparative composition in the tomato genome
Ying Wang et al. Genetics. 2006 Apr.
Abstract
Eleven sequenced BACs were annotated and localized via FISH to tomato pachytene chromosomes providing the first global insights into the compositional differences of euchromatin and pericentromeric heterochromatin in this model dicot species. The results indicate that tomato euchromatin has a gene density (6.7 kb/gene) similar to that of Arabidopsis and rice. Thus, while the euchromatin comprises only 25% of the tomato nuclear DNA, it is sufficient to account for approximately 90% of the estimated 38,000 nontransposon genes that compose the tomato genome. Moreover, euchromatic BACs were largely devoid of transposons or other repetitive elements. In contrast, BACs assigned to the pericentromeric heterochromatin had a gene density 10-100 times lower than that of the euchromatin and are heavily populated by retrotransposons preferential to the heterochromatin-the most abundant transposons belonging to the Jinling Ty3/gypsy-like retrotransposon family. Jinling elements are highly methylated and rarely transcribed. Nonetheless, they have spread throughout the pericentromeric heterochromatin in tomato and wild tomato species fairly recently-well after tomato diverged from potato and other related solanaceous species. The implications of these findings on evolution and on sequencing the genomes of tomato and other solanaceous species are discussed.
Figures
Figure 1.
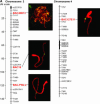
FISH with BACs located in euchromatin and heterochromatin. The distance of the FISH signals in a relative scale from centromere to telomere were correlated to the genetic mapping positions on the highly saturated tomato F2.2000 map (F
ulton
et al. 2002;
). BAC19 and 62O11 were mapped with high mapping confidence of LOD >3, and other BACs were mapped with LOD <2 as interval markers. Chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and BAC DNA was labeled with digoxigenin-16-dUTP.
Figure 1.
FISH with BACs located in euchromatin and heterochromatin. The distance of the FISH signals in a relative scale from centromere to telomere were correlated to the genetic mapping positions on the highly saturated tomato F2.2000 map (F
ulton
et al. 2002;
). BAC19 and 62O11 were mapped with high mapping confidence of LOD >3, and other BACs were mapped with LOD <2 as interval markers. Chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and BAC DNA was labeled with digoxigenin-16-dUTP.
Figure 1.
FISH with BACs located in euchromatin and heterochromatin. The distance of the FISH signals in a relative scale from centromere to telomere were correlated to the genetic mapping positions on the highly saturated tomato F2.2000 map (F
ulton
et al. 2002;
). BAC19 and 62O11 were mapped with high mapping confidence of LOD >3, and other BACs were mapped with LOD <2 as interval markers. Chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and BAC DNA was labeled with digoxigenin-16-dUTP.
Figure 2.
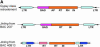
(A) Structure of a regular Ty3/Gypsy retroelement and Jinliing elements found in tomato BAC 40B13 and 2O7. (B) The structural annotation of 11 tomato BACs, indicating the putative genes and transposable elements. The number in the putative gene indicates the supporting evidence: 1, significant match to Solanaceae EST; 2, significant match to Arabidopsis gene; 3, both Solanaceae EST match and Arabidopsis match; 4, computational prediction only with no significant matches found in Solanaceae ESTs, Arabidopsis, or GenBank. The three random BACs (181K1, 181C9, and 181O9) from tomato heterochromatic regions all fell into category 1—containing no genes other than those related to transposons. The other three BACs derived from heterochromatin were assigned into categories 1 and 2 (containing both transposons and nontransposon genes). BACs from euchromatin, except 240K4, fell into category 3, containing only nontransposon genes.
Figure 2.
(A) Structure of a regular Ty3/Gypsy retroelement and Jinliing elements found in tomato BAC 40B13 and 2O7. (B) The structural annotation of 11 tomato BACs, indicating the putative genes and transposable elements. The number in the putative gene indicates the supporting evidence: 1, significant match to Solanaceae EST; 2, significant match to Arabidopsis gene; 3, both Solanaceae EST match and Arabidopsis match; 4, computational prediction only with no significant matches found in Solanaceae ESTs, Arabidopsis, or GenBank. The three random BACs (181K1, 181C9, and 181O9) from tomato heterochromatic regions all fell into category 1—containing no genes other than those related to transposons. The other three BACs derived from heterochromatin were assigned into categories 1 and 2 (containing both transposons and nontransposon genes). BACs from euchromatin, except 240K4, fell into category 3, containing only nontransposon genes.
Figure 3.
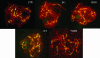
FISH with repetitive elements (LTR, RT, and IN from the Jinling element in 40B13; the GAG domain from the Jinling element in BAC 2O7; and TGRII).
Figure 4.
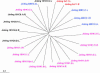
A phylogenetic tree illustrating the evolutionary relationships among Jinling elements based on the LTRs at the opposite ends. The alignment of all Jinling LTRs is in supplemental file 1 at
http://www.genetics.org/supplemental/
.
Figure 5.
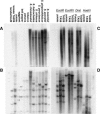
Autoradiograms derived from _Hin_dIII-digested genomic DNA of S. pimpinellifolium (LA1589), S. chmielewskii (LA1316), S. peruvianum (LA1708), S. chilense (LA1959), S. pennellii (TA56), S. neorickii (LA2133), S. Habrochaites (LA1777), S . lycopersicum (TA209), S. tuberosum (potato), Capsicum annuum (garden pepper), Petunia × hybrida hort. ex E. Vilm., and Solanum melongena (eggplant), probed with Jinling LTR (A) and 45S rDNA (R45S) (B). Lanes of TA209 and LA2133 were adjusted to display a similar amount of DNA as the other Solanum species for better comparison. (A) Potato showed weak hybridization signals, which is due to the strong background caused by overloading of potato DNA. To further test the availability of LTR sequences in potato, genomic DNAs of potato, TA209, and TA56 were digested with _Eco_RI, _Eco_RV, _Dra_I, and _Hae_III and hybridized with LTR (C) and R45S (D).
Figure 6.
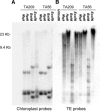
Autoradiograms derived from S . lycopersicum (TA209) and S. Pennellii (TA56) digested with _Msp_I (C*CGG), _Hpa_II (C*C*GG), _Eco_RII (CC*(A/T)GG), and _Bst_NI (CC(A/T)GG). Southern hybridizations were performed using unmethylated chloroplast genes (AY216521, AF397080, and AF263101) (A) and Jinling in 40B13 (B) as probes. The enzyme digestion will be blocked if the cytosines with asterisks are methylated.
Similar articles
- Solanum lycopersicum cv. Heinz 1706 chromosome 6: distribution and abundance of genes and retrotransposable elements.
Peters SA, Datema E, Szinay D, van Staveren MJ, Schijlen EG, van Haarst JC, Hesselink T, Abma-Henkens MH, Bai Y, de Jong H, Stiekema WJ, Klein Lankhorst RM, van Ham RC. Peters SA, et al. Plant J. 2009 Jun;58(5):857-69. doi: 10.1111/j.1365-313X.2009.03822.x. Epub 2009 Feb 4. Plant J. 2009. PMID: 19207213 - Comparative analysis of pepper and tomato reveals euchromatin expansion of pepper genome caused by differential accumulation of Ty3/Gypsy-like elements.
Park M, Jo S, Kwon JK, Park J, Ahn JH, Kim S, Lee YH, Yang TJ, Hur CG, Kang BC, Kim BD, Choi D. Park M, et al. BMC Genomics. 2011 Jan 29;12:85. doi: 10.1186/1471-2164-12-85. BMC Genomics. 2011. PMID: 21276256 Free PMC article. - High-resolution chromosome mapping of BACs using multi-colour FISH and pooled-BAC FISH as a backbone for sequencing tomato chromosome 6.
Szinay D, Chang SB, Khrustaleva L, Peters S, Schijlen E, Bai Y, Stiekema WJ, van Ham RC, de Jong H, Klein Lankhorst RM. Szinay D, et al. Plant J. 2008 Nov;56(4):627-37. doi: 10.1111/j.1365-313X.2008.03626.x. Epub 2008 Sep 18. Plant J. 2008. PMID: 18643986 - Role of fluorescence in situ hybridization in sequencing the tomato genome.
Stack SM, Royer SM, Shearer LA, Chang SB, Giovannoni JJ, Westfall DH, White RA, Anderson LK. Stack SM, et al. Cytogenet Genome Res. 2009;124(3-4):339-50. doi: 10.1159/000218137. Epub 2009 Jun 25. Cytogenet Genome Res. 2009. PMID: 19556785 Review. - An overview of plant chromosome structure.
Gill N, Hans CS, Jackson S. Gill N, et al. Cytogenet Genome Res. 2008;120(3-4):194-201. doi: 10.1159/000121067. Epub 2008 May 22. Cytogenet Genome Res. 2008. PMID: 18504347 Review. No abstract available.
Cited by
- Sequence-based SSR marker development and their application in defining the Introgressions of LA0716 (Solanum pennellii) in the background of cv. M82 (Solanum lycopersicum).
Long W, Li Y, Zhou W, Ling HQ, Zheng S. Long W, et al. PLoS One. 2013 Dec 5;8(12):e81091. doi: 10.1371/journal.pone.0081091. eCollection 2013. PLoS One. 2013. PMID: 24339899 Free PMC article. - Highly distinct chromosomal structures in cowpea (Vigna unguiculata), as revealed by molecular cytogenetic analysis.
Iwata-Otsubo A, Lin JY, Gill N, Jackson SA. Iwata-Otsubo A, et al. Chromosome Res. 2016 May;24(2):197-216. doi: 10.1007/s10577-015-9515-3. Epub 2016 Jan 12. Chromosome Res. 2016. PMID: 26758200 Free PMC article. - An interspecific linkage map of SSR and intronic polymorphism markers in tomato.
Shirasawa K, Asamizu E, Fukuoka H, Ohyama A, Sato S, Nakamura Y, Tabata S, Sasamoto S, Wada T, Kishida Y, Tsuruoka H, Fujishiro T, Yamada M, Isobe S. Shirasawa K, et al. Theor Appl Genet. 2010 Aug;121(4):731-9. doi: 10.1007/s00122-010-1344-3. Epub 2010 Apr 30. Theor Appl Genet. 2010. PMID: 20431859 Free PMC article. - Mapping pachytene chromosomes of coffee using a modified protocol for fluorescence in situ hybridization.
Iacia AA, Pinto-Maglio CA. Iacia AA, et al. AoB Plants. 2013;5:plt040. doi: 10.1093/aobpla/plt040. Epub 2013 Nov 7. AoB Plants. 2013. PMID: 24244840 Free PMC article. - Multiple QTL for horticultural traits and quantitative resistance to Phytophthora infestans linked on Solanum habrochaites chromosome 11.
Haggard JE, Johnson EB, St Clair DA. Haggard JE, et al. G3 (Bethesda). 2014 Dec 12;5(2):219-33. doi: 10.1534/g3.114.014654. G3 (Bethesda). 2014. PMID: 25504736 Free PMC article.
References
- Arumuganathan, K., J. P. Slattery, S. D. Tanksley and E. D. Earle, 1991. Preparation and flow cytometric analysis of metaphase chromosomes of tomato. Theor. Appl. Genet. 82: 101–111. - PubMed
- Bender, J., 2004. Chromatin-based silencing mechanisms. Curr. Opin. Plant Biol. 7: 521–526. - PubMed
- Bennetzen, J. L., K. Schrick, P. S. Springer, W. E. Brown and P. Sanmiguel, 1994. Active maize genes are unmodified and flanked by diverse classes of modified highly repetitive DNA. Genome 37: 565–576. - PubMed
- Bernatzky, R., and S. D. Tanksley, 1986. Majority of random cDNA clones correspond to single loci in the tomato genome. Mol. Gen. Genet. 203: 8–14.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources