Identification of distinct molecular phenotypes in acute megakaryoblastic leukemia by gene expression profiling - PubMed (original) (raw)
. 2006 Feb 28;103(9):3339-44.
doi: 10.1073/pnas.0511150103. Epub 2006 Feb 21.
Aravind Subramanian, Claudia Langebrake, Dirk Reinhardt, Olivier Bernard, Paola Ballerini, André Baruchel, Hélène Cavé, Nicole Dastugue, Henrik Hasle, Gertjan L Kaspers, Michel Lessard, Lucienne Michaux, Paresh Vyas, Elisabeth van Wering, Christian M Zwaan, Todd R Golub, Stuart H Orkin
Affiliations
- PMID: 16492768
- PMCID: PMC1413912
- DOI: 10.1073/pnas.0511150103
Identification of distinct molecular phenotypes in acute megakaryoblastic leukemia by gene expression profiling
Jean-Pierre Bourquin et al. Proc Natl Acad Sci U S A. 2006.
Abstract
Individuals with Down syndrome (DS) are predisposed to develop acute megakaryoblastic leukemia (AMKL), characterized by expression of truncated GATA1 transcription factor protein (GATA1s) due to somatic mutation. The treatment outcome for DS-AMKL is more favorable than for AMKL in non-DS patients. To gain insight into gene expression differences in AMKL, we compared 24 DS and 39 non-DS AMKL samples. We found that non-DS-AMKL samples cluster in two groups, characterized by differences in expression of HOX/TALE family members. Both of these groups are distinct from DS-AMKL, independent of chromosome 21 gene expression. To explore alterations of the GATA1 transcriptome, we used cross-species comparison with genes regulated by GATA1 expression in murine erythroid precursors. Genes repressed after GATA1 induction in the murine system, most notably GATA-2, MYC, and KIT, show increased expression in DS-AMKL, suggesting that GATA1s fail to repress this class of genes. Only a subset of genes that are up-regulated upon GATA1 induction in the murine system show increased expression in DS-AMKL, including GATA1 and BACH1, a probable negative regulator of megakaryocytic differentiation located on chromosome 21. Surprisingly, expression of the chromosome 21 gene RUNX1, a known regulator of megakaryopoiesis, was not elevated in DS-AMKL. Our results identify relevant signatures for distinct AMKL entities and provide insight into gene expression changes associated with these related leukemias.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
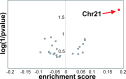
Fig. 1.
Chromosome 21 gene expression is increased in DS-AMKL. Genes were listed in sets according to their chromosomal location. Enrichment for chromosome sets was assessed by
gsea
for the comparison DS- vs. non-DS-AMKL. Positive enrichment score means higher expression in DS.
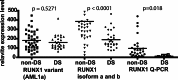
Fig. 2.
RUNX1 expression in DS-AMKL. Runx 1 levels were decreased in DS- compared with non-DS-AMKL as detected with the probe set 209360_s_at [specific for RUNX1 isoforms a and b, National Center for Biotechnology Information (NCBI) NM_001001890] and by quantitative PCR (Q-PCR) with a probe amplifying RUNX1 isoforms a and b but not the shorter variant AML1a, NCBI D43967 (probe Hs00231079_m1, Applied Biosystems). Variant AML1a detected by probe set 210365_at. Statistical analysis: two-tailed nonparametric Mann–Whitney test (GraphPad
prism 4.0
, San Diego).
Fig. 3.
Cross-species analysis with a murine GATA1 inducible system. (a) Principle of GSEA. Ortholog genes correspond to genes induced or repressed by GATA1 in the mouse system (see Table 7) are listed in sets. The human genes are rank-ordered based on differential expression for DS-AMKL vs. non-DS-AMKL.
gsea
scores the relative position of the each gene in the mouse set in the leukemia signature. (b)
gsea
results. Genes down-modulated by GATA1 in the mouse experiment are listed as they appear in the rank-ordered list of markers for DS vs. non-DS-AMKL, from top-highest to bottom-lowest relative level of expression. Enrichment score = 0.69, P = 0.029. Twenty-nine genes with significant differences between DS and non-DS are listed on the right.
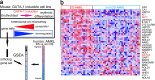
Fig. 4.
Identification of consensus cluster in non-DS-AMKL. (a) Consensus matrix produced by hierarchical clustering (K = 2). The samples are listed in the same order on the x and y axis. The intensity of the red color of the square for each sample combination corresponds to the frequency the samples cluster together in the iterations of dataset perturbation. Cluster I (CC I), 13 samples; Cluster II (CC II), 23 samples. (b) Expression profiles of the two AMKL clusters. The top 60 genes associated with each AMKL clusters are shown. Color scale at bottom indicates relative expression to the median. Red, high-level expression; blue, low-level expression. (c) List of selected genes (complete data in Table 8, which is published as supporting information on the PNAS web site).
Similar articles
- Physical association of the patient-specific GATA1 mutants with RUNX1 in acute megakaryoblastic leukemia accompanying Down syndrome.
Xu G, Kanezaki R, Toki T, Watanabe S, Takahashi Y, Terui K, Kitabayashi I, Ito E. Xu G, et al. Leukemia. 2006 Jun;20(6):1002-8. doi: 10.1038/sj.leu.2404223. Leukemia. 2006. PMID: 16628190 - Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome.
Wechsler J, Greene M, McDevitt MA, Anastasi J, Karp JE, Le Beau MM, Crispino JD. Wechsler J, et al. Nat Genet. 2002 Sep;32(1):148-52. doi: 10.1038/ng955. Epub 2002 Aug 12. Nat Genet. 2002. PMID: 12172547 - Mutations in exon 2 of GATA1 are early events in megakaryocytic malignancies associated with trisomy 21.
Rainis L, Bercovich D, Strehl S, Teigler-Schlegel A, Stark B, Trka J, Amariglio N, Biondi A, Muler I, Rechavi G, Kempski H, Haas OA, Izraeli S. Rainis L, et al. Blood. 2003 Aug 1;102(3):981-6. doi: 10.1182/blood-2002-11-3599. Epub 2003 Mar 20. Blood. 2003. PMID: 12649131 - Acute megakaryoblastic leukaemia (AMKL) and transient myeloproliferative disorder (TMD) in Down syndrome: a multi-step model of myeloid leukaemogenesis.
Roy A, Roberts I, Norton A, Vyas P. Roy A, et al. Br J Haematol. 2009 Oct;147(1):3-12. doi: 10.1111/j.1365-2141.2009.07789.x. Epub 2009 Jul 6. Br J Haematol. 2009. PMID: 19594743 Review. - Down myeloid disorders: a paradigm for childhood preleukaemia and leukaemia and insights into normal megakaryopoiesis.
Vyas P, Roberts I. Vyas P, et al. Early Hum Dev. 2006 Dec;82(12):767-73. doi: 10.1016/j.earlhumdev.2006.09.016. Epub 2006 Oct 24. Early Hum Dev. 2006. PMID: 17064858 Review.
Cited by
- Differential sensitivities of transcription factor target genes underlie cell type-specific gene expression profiles.
Johnson KD, Kim SI, Bresnick EH. Johnson KD, et al. Proc Natl Acad Sci U S A. 2006 Oct 24;103(43):15939-44. doi: 10.1073/pnas.0604041103. Epub 2006 Oct 16. Proc Natl Acad Sci U S A. 2006. PMID: 17043224 Free PMC article. - Development of acute megakaryoblastic leukemia in Down syndrome is associated with sequential epigenetic changes.
Malinge S, Chlon T, Doré LC, Ketterling RP, Tallman MS, Paietta E, Gamis AS, Taub JW, Chou ST, Weiss MJ, Crispino JD, Figueroa ME. Malinge S, et al. Blood. 2013 Oct 3;122(14):e33-43. doi: 10.1182/blood-2013-05-503011. Epub 2013 Aug 26. Blood. 2013. PMID: 23980066 Free PMC article. - Oncogenic Gata1 causes stage-specific megakaryocyte differentiation delay.
Juban G, Sakakini N, Chagraoui H, Cruz Hernandez D, Cheng Q, Soady K, Stoilova B, Garnett C, Waithe D, Otto G, Doondeea J, Usukhbayar B, Karkoulia E, Alexiou M, Strouboulis J, Morrissey E, Roberts I, Porcher C, Vyas P. Juban G, et al. Haematologica. 2021 Apr 1;106(4):1106-1119. doi: 10.3324/haematol.2019.244541. Haematologica. 2021. PMID: 32527952 Free PMC article. - RUNX1 isoform disequilibrium promotes the development of trisomy 21-associated myeloid leukemia.
Gialesaki S, Bräuer-Hartmann D, Issa H, Bhayadia R, Alejo-Valle O, Verboon L, Schmell AL, Laszig S, Regényi E, Schuschel K, Labuhn M, Ng M, Winkler R, Ihling C, Sinz A, Glaß M, Hüttelmaier S, Matzk S, Schmid L, Strüwe FJ, Kadel SK, Reinhardt D, Yaspo ML, Heckl D, Klusmann JH. Gialesaki S, et al. Blood. 2023 Mar 9;141(10):1105-1118. doi: 10.1182/blood.2022017619. Blood. 2023. PMID: 36493345 Free PMC article. - RUNX1 Upregulation Causes Mitochondrial Dysfunction via Regulating the PI3K-Akt Pathway in iPSC from Patients with Down Syndrome.
Liu Y, Zhang Y, Ren Z, Zeng F, Yan J. Liu Y, et al. Mol Cells. 2023 Apr 30;46(4):219-230. doi: 10.14348/molcells.2023.2095. Epub 2023 Jan 10. Mol Cells. 2023. PMID: 36625318 Free PMC article.
References
- Athale U. H., Razzouk B. I., Raimondi S. C., Tong X., Behm F. G., Head D. R., Srivastava D. K., Rubnitz J. E., Bowman L., Pui C. H., Ribeiro R. C. Blood. 2001;97:3727–3732. - PubMed
- Creutzig U., Reinhardt D., Diekamp S., Dworzak M., Stary J., Zimmermann M. Leukemia. 2005;19:1355–1360. - PubMed
- Dastugue N., Lafage-Pochitaloff M., Pages M. P., Radford I., Bastard C., Talmant P., Mozziconacci M. J., Leonard C., Bilhou-Nabera C., Cabrol C., et al. Blood. 2002;100:618–626. - PubMed
- Ma Z., Morris S. W., Valentine V., Li M., Herbrick J. A., Cui X., Bouman D., Li Y., Mehta P. K., Nizetic D., Kaneko Y., et al. Nat. Genet. 2001;28:220–221. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous