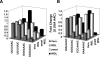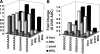Comparative study of the effects of heptameric slippery site composition on -1 frameshifting among different eukaryotic systems - PubMed (original) (raw)
Comparative Study
Comparative study of the effects of heptameric slippery site composition on -1 frameshifting among different eukaryotic systems
Ewan P Plant et al. RNA. 2006 Apr.
Abstract
Studies of programmed -1 ribosomal frameshifting (-1 PRF) have been approached over the past two decades by many different laboratories using a diverse array of virus-derived frameshift signals in translational assay systems derived from a variety of sources. Though it is generally acknowledged that both absolute and relative -1 PRF efficiency can vary in an assay system-dependent manner, no methodical study of this phenomenon has been undertaken. To address this issue, a series of slippery site mutants of the SARS-associated coronavirus frameshift signal were systematically assayed in four different eukaryotic translational systems. HIV-1 promoted frameshifting was also compared between Escherichia coli and a human T-cell line expression systems. The results of these analyses highlight different aspects of each system, suggesting in general that (1) differences can be due to the assay systems themselves; (2) phylogenetic differences in ribosome structure can affect frameshifting efficiency; and (3) care must be taken to employ the closest phylogenetic match between a specific -1 PRF signal and the choice of translational assay system.
Figures
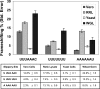
FIGURE 1.
Frameshifting efficiency from individual slippery sites varies depending on the translational assay system. Luminescence from test constructs is expressed as a percentage of the nonframe-shifting control for different slippery sites. The shading of the bars represents frameshifting stimulated by different translational systems: dark gray for Vero cells, light gray for reticulocyte lysate, white for yeast cells, and black for wheat germ lysate. Percentages of frameshifting and standard errors are indicated in both the figure and the table below.
FIGURE 2.
Influence of the A- and the P-site codons on −1 frame-shifting in different translational systems. (A) Influence of the A-site codon. (B) Influence of the P-site codon. Frameshifting efficiencies were determined and expressed as fold change of the wild-type sequence (U UUA AAC) for each construct. The shading of the bars is the same as in Figure 1 ▶. _P_-values for fold change are in Table 1 ▶.
FIGURE 3.
Effects of the seventh position of the slippery site on −1 frameshifting in different translational systems. (A) Effects with a SARS-like slippery site. (B) Effects with a (L–A)-like slippery site. Frameshifting efficiencies were determined and expressed as fold change of the SARS wild-type sequence (U UUA AAC) for each construct. The shading of the bars is the same as in Figure 1 ▶. _P_-values for fold change are in Table 1 ▶.
Similar articles
- Analyses of frameshifting at UUU-pyrimidine sites.
Schwartz R, Curran JF. Schwartz R, et al. Nucleic Acids Res. 1997 May 15;25(10):2005-11. doi: 10.1093/nar/25.10.2005. Nucleic Acids Res. 1997. PMID: 9115369 Free PMC article. - Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting.
Hansen TM, Reihani SN, Oddershede LB, Sørensen MA. Hansen TM, et al. Proc Natl Acad Sci U S A. 2007 Apr 3;104(14):5830-5. doi: 10.1073/pnas.0608668104. Epub 2007 Mar 27. Proc Natl Acad Sci U S A. 2007. PMID: 17389398 Free PMC article. - The three transfer RNAs occupying the A, P and E sites on the ribosome are involved in viral programmed -1 ribosomal frameshift.
Léger M, Dulude D, Steinberg SV, Brakier-Gingras L. Léger M, et al. Nucleic Acids Res. 2007;35(16):5581-92. doi: 10.1093/nar/gkm578. Epub 2007 Aug 17. Nucleic Acids Res. 2007. PMID: 17704133 Free PMC article. - Targeting Ribosomal Frameshifting as an Antiviral Strategy: From HIV-1 to SARS-CoV-2.
Anokhina VS, Miller BL. Anokhina VS, et al. Acc Chem Res. 2021 Sep 7;54(17):3349-3361. doi: 10.1021/acs.accounts.1c00316. Epub 2021 Aug 17. Acc Chem Res. 2021. PMID: 34403258 Review. - Slippy-Sloppy translation: a tale of programmed and induced-ribosomal frameshifting.
Champagne J, Mordente K, Nagel R, Agami R. Champagne J, et al. Trends Genet. 2022 Nov;38(11):1123-1133. doi: 10.1016/j.tig.2022.05.009. Epub 2022 May 28. Trends Genet. 2022. PMID: 35641342 Review.
Cited by
- Ribosomal frameshifting at normal codon repeats recodes functional chimeric proteins in human.
Ren G, Gu X, Zhang L, Gong S, Song S, Chen S, Chen Z, Wang X, Li Z, Zhou Y, Li L, Yang J, Lai F, Dang Y. Ren G, et al. Nucleic Acids Res. 2024 Mar 21;52(5):2463-2479. doi: 10.1093/nar/gkae035. Nucleic Acids Res. 2024. PMID: 38281188 Free PMC article. - Viral and Cellular mRNA Translation in Coronavirus-Infected Cells.
Nakagawa K, Lokugamage KG, Makino S. Nakagawa K, et al. Adv Virus Res. 2016;96:165-192. doi: 10.1016/bs.aivir.2016.08.001. Epub 2016 Sep 10. Adv Virus Res. 2016. PMID: 27712623 Free PMC article. Review. - Virus versus host cell translation love and hate stories.
Komarova AV, Haenni AL, Ramírez BC. Komarova AV, et al. Adv Virus Res. 2009;73:99-170. doi: 10.1016/S0065-3527(09)73003-9. Adv Virus Res. 2009. PMID: 19695382 Free PMC article. Review. - The highly conserved codon following the slippery sequence supports -1 frameshift efficiency at the HIV-1 frameshift site.
Mathew SF, Crowe-McAuliffe C, Graves R, Cardno TS, McKinney C, Poole ES, Tate WP. Mathew SF, et al. PLoS One. 2015 Mar 25;10(3):e0122176. doi: 10.1371/journal.pone.0122176. eCollection 2015. PLoS One. 2015. PMID: 25807539 Free PMC article. - The role of programmed-1 ribosomal frameshifting in coronavirus propagation.
Plant EP, Dinman JD. Plant EP, et al. Front Biosci. 2008 May 1;13:4873-81. doi: 10.2741/3046. Front Biosci. 2008. PMID: 18508552 Free PMC article. Review.
References
- Barak, Z., Gallant, J., Lindsley, D., Kwieciszewki, B., and Heidel, D. 1996. Enhanced ribosome frameshifting in stationary phase cells. J. Mol. Biol. 263: 140–148. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous