The use of biotin tagging in Saccharomyces cerevisiae improves the sensitivity of chromatin immunoprecipitation - PubMed (original) (raw)
The use of biotin tagging in Saccharomyces cerevisiae improves the sensitivity of chromatin immunoprecipitation
Folkert J van Werven et al. Nucleic Acids Res. 2006.
Abstract
Affinity tagging has been used in many global studies towards protein function. We describe a highly efficient system for in vivo biotinylation of transcription factors in the yeast Saccharomyces cerevisiae, which is based on the bacterial BirA biotin ligase. The strength of the biotin-streptavidin interaction was exploited to improve detection of in vivo protein-DNA complexes in chromatin immunoprecipitation (ChIP) experiments. In a test system using the biotin-tagged LexA DNA-binding protein, we found that stringent washing conditions resulted in a strong improvement of the signal-to-noise ratios. Yeast strains with chromosomally integrated versions of tagged transcription factor genes were generated using N- or C-terminal biotin-tagging cassettes. ChIP experiments with biotinylated Rbp3p, a RNA polymerase II subunit, showed that Rbp3p-binding could even be detected at weakly expressed genes. Other methods failed to detect RNA polymerase II binding at such genes. Our results show that biotinylation of yeast transcription factors improves the detection of in vivo protein-DNA complexes.
Figures
Figure 1
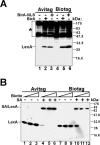
Comparison of biotinylation effiencies of LexA proteins by BirA or BirA-NLS. (A) LexA-Avitag or LexA-Biotag proteins were coexpressed with either BirA or BirA-NLS proteins as indicated. Biotinylated proteins were detected with SA-HRP as the primary detection agent. Endogenously biotinylated proteins are indicated by the asterisk. (B) Yeast cells were grown in the presence of increasing concentrations of biotin. Protein extracts were incubated with streptavidin (SA) as indicated, and immunoblots were developed with antibodies recognizing LexA protein.
Figure 2
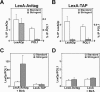
ChIP analysis of LexA binding to chromosomal LexOp sequences. Chromatin was extracted from crosslinked EGY48 cells expressing the indicated LexA proteins. Captured protein–DNA complexes were washed under standard or stringent conditions (containing 3% SDS). (A and B) ChIP analysis of LexA-Avitag and LexA-TAP binding. The bound material was analyzed by LexAOp and POL1 primer sets, and the signals are expressed relative to the input. Please note the difference in scale of the specific signal to the left and the non-binding control to the right of the figure. (C) Similar to (A and B) except that enrichment over the POL1 control is indicated. (D) Control ChIP analysis of LexA-Avitag proteins in the absence of BirA expression (left columns) and of non-tagged LexA protein in the presence of BirA expression (right columns). ChIP analysis was performed as shown in (A) and the signals are expressed relative to the POL1 control sequences.
Figure 3
Tools for gene tagging by chromosomal integration. (A) Plasmids for PCR-based insertion of Avitag sequences at the C- and N- terminus of ORFs. The position of the upstream and downstream primers described in (B) are indicated. (B) Universal primer sequences for the tagging cassettes depicted in (A). To each primer 50 nt of yeast genomic sequence flanking the integration site should be fused. The reading frames are indicated for the appropriate primer sequences. (C) Plasmid map of pRS313-BirA-NLS. This represents a low-copy BirA-NLS expression plasmid bearing a HIS3 marker gene. The BirA-NLS expression cassette has also been inserted in low-copy plasmids bearing TRP1 (pRS314-BirA-NLS), LEU2 (pRS315-BirA-NLS), URA3 (pRS316-BirA-NLS).
Figure 4
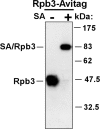
Biotinylation efficiency of Rpb3-Avitag. Protein extract of Rpb3-Avitag strain was incubated with SA as indicated. Rpb3p was detected by immunoblotting using antibodies specific for the co-integrated Myc epitope.
Figure 5
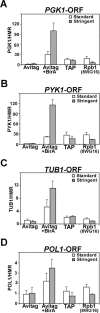
ChIP analysis of Rpb3-Avitag, Rpb3-TAP and Rpb1 (using 8WG16 antibodies) binding to the ORFs of PGK1 (A), PYK1 (B), TUB1 (C) and POL1 (D). Chromatin extracts of crosslinked cells were prepared and association to the indicated gene fragments were analyzed in ChIP. Captured protein–DNA complexes were washed under standard or stringent conditions (containing 3% SDS). Binding to tested regions is represented as enrichment over the HMR silent mating-type locus.
Similar articles
- An immobilized biotin ligase: surface display of Escherichia coli BirA on Saccharomyces cerevisiae.
Parthasarathy R, Bajaj J, Boder ET. Parthasarathy R, et al. Biotechnol Prog. 2005 Nov-Dec;21(6):1627-31. doi: 10.1021/bp050279t. Biotechnol Prog. 2005. PMID: 16321044 - In vivo biotinylation of the major histocompatibility complex (MHC) class II/peptide complex by coexpression of BirA enzyme for the generation of MHC class II/tetramers.
Yang J, Jaramillo A, Shi R, Kwok WW, Mohanakumar T. Yang J, et al. Hum Immunol. 2004 Jul;65(7):692-9. doi: 10.1016/j.humimm.2004.04.001. Hum Immunol. 2004. PMID: 15301857 - Isolation of transcription factor complexes by in vivo biotinylation tagging and direct binding to streptavidin beads.
Rodriguez P, Braun H, Kolodziej KE, de Boer E, Campbell J, Bonte E, Grosveld F, Philipsen S, Strouboulis J. Rodriguez P, et al. Methods Mol Biol. 2006;338:305-23. doi: 10.1385/1-59745-097-9:305. Methods Mol Biol. 2006. PMID: 16888367 - Isolation and characterization of hematopoietic transcription factor complexes by in vivo biotinylation tagging and mass spectrometry.
Grosveld F, Rodriguez P, Meier N, Krpic S, Pourfarzad F, Papadopoulos P, Kolodziej K, Patrinos GP, Hostert A, Strouboulis J. Grosveld F, et al. Ann N Y Acad Sci. 2005;1054:55-67. doi: 10.1196/annals.1345.008. Ann N Y Acad Sci. 2005. PMID: 16339652 Review. - DNA protein interactions at the rRNA of Saccharomyces cerevisiae.
Cioci F, Di Felice F, Chiani F, Camilloni G. Cioci F, et al. Ital J Biochem. 2007 Jun;56(2):81-90. Ital J Biochem. 2007. PMID: 17722648 Review.
Cited by
- Optimal use of tandem biotin and V5 tags in ChIP assays.
Kolodziej KE, Pourfarzad F, de Boer E, Krpic S, Grosveld F, Strouboulis J. Kolodziej KE, et al. BMC Mol Biol. 2009 Feb 5;10:6. doi: 10.1186/1471-2199-10-6. BMC Mol Biol. 2009. PMID: 19196479 Free PMC article. - Nonradioactive direct telomerase activity detection using biotin-labeled primers.
Wang R, Li J, Jin R, Ye Q, Cheng L, Liu R. Wang R, et al. J Clin Lab Anal. 2021 Jun;35(6):e23800. doi: 10.1002/jcla.23800. Epub 2021 May 7. J Clin Lab Anal. 2021. PMID: 33960443 Free PMC article. - Mutation of a phosphorylatable residue in Put3p affects the magnitude of rapamycin-induced PUT1 activation in a Gat1p-dependent manner.
Leverentz MK, Campbell RN, Connolly Y, Whetton AD, Reece RJ. Leverentz MK, et al. J Biol Chem. 2009 Sep 4;284(36):24115-22. doi: 10.1074/jbc.M109.030361. Epub 2009 Jul 1. J Biol Chem. 2009. PMID: 19574222 Free PMC article. - The Scc2/Scc4 cohesin loader determines the distribution of cohesin on budding yeast chromosomes.
Kogut I, Wang J, Guacci V, Mistry RK, Megee PC. Kogut I, et al. Genes Dev. 2009 Oct 1;23(19):2345-57. doi: 10.1101/gad.1819409. Genes Dev. 2009. PMID: 19797771 Free PMC article. - Improved genome-wide localization by ChIP-chip using double-round T7 RNA polymerase-based amplification.
van Bakel H, van Werven FJ, Radonjic M, Brok MO, van Leenen D, Holstege FC, Timmers HT. van Bakel H, et al. Nucleic Acids Res. 2008 Mar;36(4):e21. doi: 10.1093/nar/gkm1144. Epub 2008 Jan 7. Nucleic Acids Res. 2008. PMID: 18180247 Free PMC article.
References
- Hassan A.H., Neely K.E., Vignali M., Reese J.C., Workman J.L. Promoter targeting of chromatin-modifying complexes. Front Biosci. 2001;6:D1054–D1064. - PubMed
- Martinez E. Multi-protein complexes in eukaryotic gene transcription. Plant Mol. Biol. 2002;50:925–947. - PubMed
- Roeder R.G. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. - PubMed
- Schaffner W. Gene regulation. A hit-and-run mechanism for transcriptional activation? Nature. 1988;336:427–428. - PubMed
- Truss M., Chalepakis G., Beato M. Interplay of steroid hormone receptors and transcription factors on the mouse mammary tumor virus promoter. J. Steroid Biochem. Mol. Biol. 1992;43:365–378. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases