Accuracy of DNA methylation pattern preservation by the Dnmt1 methyltransferase - PubMed (original) (raw)
Accuracy of DNA methylation pattern preservation by the Dnmt1 methyltransferase
Rachna Goyal et al. Nucleic Acids Res. 2006.
Abstract
DNA methyltransferase 1 (Dnmt1) has a central role in copying the pattern of DNA methylation after replication which is one manifestation of epigenetic inheritance. With oligonculeotide substrates we show that mouse Dnmt1 has a 30- to 40-fold preference for hemimethylated DNA that is almost lost after addition of fully methylated oligonucleotides. Using long hemimethylated DNA substrates that carry defined methylation patterns and bisulfite analysis of the methylation reaction products, we show a 15-fold preference for hemimethylated CG sites. Dnmt1 moves along the DNA in a random walk methylating hemimethylated substrates with high processivity (>50 sites are visited on average which corresponds to linear diffusion over 6000 bp). The frequency of skipping sites is very low (<0.3%) and there is no detectable flanking sequence preference. CGCTC sites tend to terminate the processive methylation of DNA by Dnmt1. Unmethylated DNA is modified non-processively with a preference for methylation at CCGG sites. We simulate the propagation of methylation patterns using a stochastic model with the specificity of Dnmt1 observed here and conclude that either methylation of several sites is required to propagate the methylation information over several cellular generations or additional epigenetic information must be used.
Figures
Figure 1
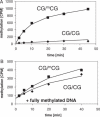
Methylation of unmethylated and hemimethylated oligonucleotides by Dnmt1. Methylation was analyzed by incorporation of radioactive methyl groups into the DNA.In (A) 1 µM of hemimethylated and unmethylated oligonucleotides (CG30hm and CG30um) and 100 nM enzyme were used. In (B) 1 µM fully methylated oligonucleotide (CG30fm) of the same sequence was added.
Figure 2
Methylation of unmethylated and hemimethylated long substrates in different reaction tubes. (A) Methylation of unmethylated substrate 1 by Dnmt1. In (B) substrate 1 was prepared in hemimethylated state at all CG sites and used for methylation by Dnmt1. Methylation was analyzed after bisulfite modification of the DNA, PCR amplification, cloning and sequencing of at least 20 individual clones for each DNA strand. In the figure, some examples of the clones obtained and sequenced are shown. Methylated CG sites are shown black, unmethylated white.
Figure 3
Methylation of unmethylated and hemimethylated substrate in one reaction. (A) Methylation of unmethylated substrate 1 and hemimethylated substrate 2 by Dnmt1 in one reaction tube. Substrate 2 is hemimethylated at all CG sites in this experiment. (B) Methylation of substrate 2 hemimethylated at all CCGG sites. Methylation was analyzed after bisulfite modification of the DNA, PCR amplification, cloning and sequencing of at least 20 individual clones for each DNA strand. In the figure, some examples of the clones obtained and sequenced are shown. Methylated CG sites are shown black, unmethylated white. In (B) the hemimethylated CCGG sites are shaded gray.
Figure 4
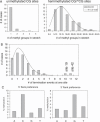
(A) Processivity of the methylation of the unmethylated and hemimethylated CG sites by Dnmt1. The figure displays the distribution of lengths of all stretches of methylated sites for the unmethylated (left) and hemimethylated substrate (right). The line in the right panel indicates a fit to the random walk model with a _P_dif of 54. (B) Distribution of the number of termination events of processive methylation of the DNA at the various CG sites. The distribution is roughly fitted by a binomial distribution of the corresponding parameters as indicated by the line, if the two sites 19 and 42 are disregarded. (C) Flanking sequence preference for methylation of unmethylated DNA by Dnmt1.
Figure 5
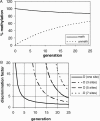
Simulation of the average methylation level of an unmethylated and hemimethylated population of DNA molecules during several generations. For the simulation an average level of maintenance methylation of 95% and a 20-fold preference for hemimethylated DNA was used. In (A) the average methylation levels of the hemimethylated and unmethylated populations at one CG site are shown, in (B) the discrimination factor (defined as the ratio of the average methylation levels of both populations) is shown if one, three, five or seven CG sites are concerned.
Similar articles
- Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1.
Valinluck V, Sowers LC. Valinluck V, et al. Cancer Res. 2007 Feb 1;67(3):946-50. doi: 10.1158/0008-5472.CAN-06-3123. Cancer Res. 2007. PMID: 17283125 - Ras regulation of DNA-methylation and cancer.
Patra SK. Patra SK. Exp Cell Res. 2008 Apr 1;314(6):1193-201. doi: 10.1016/j.yexcr.2008.01.012. Epub 2008 Jan 26. Exp Cell Res. 2008. PMID: 18282569 Review. - Cell and molecular biology of DNA methyltransferase 1.
Mohan KN, Chaillet JR. Mohan KN, et al. Int Rev Cell Mol Biol. 2013;306:1-42. doi: 10.1016/B978-0-12-407694-5.00001-8. Int Rev Cell Mol Biol. 2013. PMID: 24016522 Review.
Cited by
- The influence of cis-regulatory elements on DNA methylation fidelity.
Teng M, Balch C, Liu Y, Li M, Huang TH, Wang Y, Nephew KP, Li L. Teng M, et al. PLoS One. 2012;7(3):e32928. doi: 10.1371/journal.pone.0032928. Epub 2012 Mar 6. PLoS One. 2012. PMID: 22412954 Free PMC article. - Epigenetic Alterations in Podocytes in Diabetic Nephropathy.
Sugita E, Hayashi K, Hishikawa A, Itoh H. Sugita E, et al. Front Pharmacol. 2021 Sep 24;12:759299. doi: 10.3389/fphar.2021.759299. eCollection 2021. Front Pharmacol. 2021. PMID: 34630127 Free PMC article. Review. - The DNMT1 inhibitor GSK-3484862 mediates global demethylation in murine embryonic stem cells.
Azevedo Portilho N, Saini D, Hossain I, Sirois J, Moraes C, Pastor WA. Azevedo Portilho N, et al. Epigenetics Chromatin. 2021 Dec 15;14(1):56. doi: 10.1186/s13072-021-00429-0. Epigenetics Chromatin. 2021. PMID: 34906184 Free PMC article. - Linking DNA Damage and Age-Related Promoter DNA Hyper-Methylation in the Intestine.
Thalheim T, Herberg M, Galle J. Thalheim T, et al. Genes (Basel). 2018 Jan 5;9(1):17. doi: 10.3390/genes9010017. Genes (Basel). 2018. PMID: 29303998 Free PMC article. - Genome-wide quantitative assessment of variation in DNA methylation patterns.
Xie H, Wang M, de Andrade A, Bonaldo Mde F, Galat V, Arndt K, Rajaram V, Goldman S, Tomita T, Soares MB. Xie H, et al. Nucleic Acids Res. 2011 May;39(10):4099-108. doi: 10.1093/nar/gkr017. Epub 2011 Jan 28. Nucleic Acids Res. 2011. PMID: 21278160 Free PMC article.
References
- Egger G., Liang G., Aparicio A., Jones P.A. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. - PubMed
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
- Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem. 2002;3:274–293. - PubMed
- Jones P.A., Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. - PubMed
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nature Rev. Genet. 2002;3:662–673. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases