Global suppression of the host antiviral response by Ebola- and Marburgviruses: increased antagonism of the type I interferon response is associated with enhanced virulence - PubMed (original) (raw)
Global suppression of the host antiviral response by Ebola- and Marburgviruses: increased antagonism of the type I interferon response is associated with enhanced virulence
John C Kash et al. J Virol. 2006 Mar.
Abstract
We studied the effect of filovirus infection on host cell gene expression by characterizing the regulation of gene expression responses in human liver cells infected with Zaire Ebolavirus (ZEBOV), Reston Ebolavirus (REBOV), and Marburgvirus (MARV), using transcriptional profiling and bioinformatics. Expression microarray analysis demonstrated that filovirus infection resulted in the up-regulation of immune-related genes and the down-regulation of many coagulation and acute-phase proteins. These studies further revealed that a common feature of filovirus virulence is suppression of key cellular antiviral responses, including TLR-, interferon (IFN) regulatory factor 3-, and PKR-related pathways. We further showed that ZEBOV and MARV were more potent antagonists of the IFN response and inhibited the expression of most of the IFN-stimulated genes (ISGs) observed in mock-infected IFN-alpha-2b treated cells, compared to REBOV infection, which activated more than 20% of these ISGs. Finally, we examined IFN-related gene expression in filovirus-infected cells treated with IFN-alpha-2b. These experiments revealed that a majority of genes induced in mock-infected cells treated with type I IFN were antagonized in treated ZEBOV- and MARV-infected cells, while in contrast, REBOV infection resulted in a significant increase in ISG expression. Analysis of STAT1 and -2 phosphorylation following IFN treatment showed a significant reduction of STAT phosphorylation for MARV but not for ZEBOV and REBOV, indicating that different mechanisms might be involved in antagonizing IFN signaling pathways by the different filovirus species. Taken together, these studies showed a correlation between antagonism of type I IFN responses and filovirus virulence.
Figures
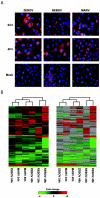
FIG. 1.
Global analysis of EBOV and MARV infection kinetics. Panel A, immunofluorescence analysis of Huh7 cells infected with ZEBOV, REBOV, or MARV at an MOI of 0.1 at 24 h and 48 h p.i. Panel B, two-dimensional agglomerative cluster matrix of genes that showed a ≥2-fold (n = 4; P < 0.01) change in expression in a least one experiment. In the left panel, genes shown in red were up-regulated and genes shown in green were down-regulated relative to uninfected Huh7 cells, while black indicates no change in gene expression. In the right panel, genes whose regulation showed a _P_ value of >0.01 are shown in gray.
FIG.2.
Control of host cell gene expression following EBOV and MARV infection. Panel A, Venn diagram showing the segregation of infection-regulated genes that showed a ≥2-fold (n = 4; P < 0.01) change in expression at 24 h p.i. Hierarchical clustering matrix of the expression of genes in set I that were preferentially regulated by ZEBOV at 24 h, in set II that were commonly regulated by ZEBOV and MARV, in set III that were common to all viral infections, and in set IV that were restricted to REBOV. Panel B, Venn diagram showing the segregation of infection-regulated genes that showed a ≥2-fold (_n_ = 4; _P_ < 0.01) change in expression at 48 h p.i. and the associated hierarchical clustering diagrams of genes in sets I to IV. Panel C, matrix showing the expression of selected inflammation- and apoptosis-related genes in ZEBOV-, REBOV-, and MARV-infected cells at 24 and 48 h p.i. In the left panel, genes shown in red were up-regulated and genes shown in green were down-regulated relative to uninfected Huh7 cells, while black indicates no change in gene expression. In the right panel, genes whose regulation showed a _P_ value of >0.01 are shown in gray. Panel D, bar graphs comparing the expression of selected mRNAs as measured by quantitative real-time PCR and cDNA expression microarray analysis. The results are presented as the log ratio of mRNA abundance in infected relative to mock-infected cells.
FIG.2.
Control of host cell gene expression following EBOV and MARV infection. Panel A, Venn diagram showing the segregation of infection-regulated genes that showed a ≥2-fold (n = 4; P < 0.01) change in expression at 24 h p.i. Hierarchical clustering matrix of the expression of genes in set I that were preferentially regulated by ZEBOV at 24 h, in set II that were commonly regulated by ZEBOV and MARV, in set III that were common to all viral infections, and in set IV that were restricted to REBOV. Panel B, Venn diagram showing the segregation of infection-regulated genes that showed a ≥2-fold (_n_ = 4; _P_ < 0.01) change in expression at 48 h p.i. and the associated hierarchical clustering diagrams of genes in sets I to IV. Panel C, matrix showing the expression of selected inflammation- and apoptosis-related genes in ZEBOV-, REBOV-, and MARV-infected cells at 24 and 48 h p.i. In the left panel, genes shown in red were up-regulated and genes shown in green were down-regulated relative to uninfected Huh7 cells, while black indicates no change in gene expression. In the right panel, genes whose regulation showed a _P_ value of >0.01 are shown in gray. Panel D, bar graphs comparing the expression of selected mRNAs as measured by quantitative real-time PCR and cDNA expression microarray analysis. The results are presented as the log ratio of mRNA abundance in infected relative to mock-infected cells.
FIG.2.
Control of host cell gene expression following EBOV and MARV infection. Panel A, Venn diagram showing the segregation of infection-regulated genes that showed a ≥2-fold (n = 4; P < 0.01) change in expression at 24 h p.i. Hierarchical clustering matrix of the expression of genes in set I that were preferentially regulated by ZEBOV at 24 h, in set II that were commonly regulated by ZEBOV and MARV, in set III that were common to all viral infections, and in set IV that were restricted to REBOV. Panel B, Venn diagram showing the segregation of infection-regulated genes that showed a ≥2-fold (_n_ = 4; _P_ < 0.01) change in expression at 48 h p.i. and the associated hierarchical clustering diagrams of genes in sets I to IV. Panel C, matrix showing the expression of selected inflammation- and apoptosis-related genes in ZEBOV-, REBOV-, and MARV-infected cells at 24 and 48 h p.i. In the left panel, genes shown in red were up-regulated and genes shown in green were down-regulated relative to uninfected Huh7 cells, while black indicates no change in gene expression. In the right panel, genes whose regulation showed a _P_ value of >0.01 are shown in gray. Panel D, bar graphs comparing the expression of selected mRNAs as measured by quantitative real-time PCR and cDNA expression microarray analysis. The results are presented as the log ratio of mRNA abundance in infected relative to mock-infected cells.
FIG.2.
Control of host cell gene expression following EBOV and MARV infection. Panel A, Venn diagram showing the segregation of infection-regulated genes that showed a ≥2-fold (n = 4; P < 0.01) change in expression at 24 h p.i. Hierarchical clustering matrix of the expression of genes in set I that were preferentially regulated by ZEBOV at 24 h, in set II that were commonly regulated by ZEBOV and MARV, in set III that were common to all viral infections, and in set IV that were restricted to REBOV. Panel B, Venn diagram showing the segregation of infection-regulated genes that showed a ≥2-fold (_n_ = 4; _P_ < 0.01) change in expression at 48 h p.i. and the associated hierarchical clustering diagrams of genes in sets I to IV. Panel C, matrix showing the expression of selected inflammation- and apoptosis-related genes in ZEBOV-, REBOV-, and MARV-infected cells at 24 and 48 h p.i. In the left panel, genes shown in red were up-regulated and genes shown in green were down-regulated relative to uninfected Huh7 cells, while black indicates no change in gene expression. In the right panel, genes whose regulation showed a _P_ value of >0.01 are shown in gray. Panel D, bar graphs comparing the expression of selected mRNAs as measured by quantitative real-time PCR and cDNA expression microarray analysis. The results are presented as the log ratio of mRNA abundance in infected relative to mock-infected cells.
FIG.3.
Increased activation and expression of IFN-regulated genes by REBOV compared to ZEBOV and MARV. Panel A, scatter plots of the expression patterns of individual genes that were ≥2-fold (n = 4; P < 0.01) up-regulated by treatment with 100 IU/ml IFN-α-2b in mock-infected cells compared to their expression in ZEBOV-, REBOV-, and MARV-infected cells at 48 h. Panel B, hierarchical clustering matrix of the ISGs that were up-regulated at 24 h and 48 h p.i. with REBOV, ZEBOV, or MARV relative to mock-infected cells treated with IFN-α-2b. In the left panel, genes shown in red were up-regulated and genes shown in green were down-regulated relative to uninfected Huh7 cells, while black indicates no change in gene expression. In the right panel, genes whose regulation showed a _P_ value of >0.01 are shown in gray.
FIG.3.
Increased activation and expression of IFN-regulated genes by REBOV compared to ZEBOV and MARV. Panel A, scatter plots of the expression patterns of individual genes that were ≥2-fold (n = 4; P < 0.01) up-regulated by treatment with 100 IU/ml IFN-α-2b in mock-infected cells compared to their expression in ZEBOV-, REBOV-, and MARV-infected cells at 48 h. Panel B, hierarchical clustering matrix of the ISGs that were up-regulated at 24 h and 48 h p.i. with REBOV, ZEBOV, or MARV relative to mock-infected cells treated with IFN-α-2b. In the left panel, genes shown in red were up-regulated and genes shown in green were down-regulated relative to uninfected Huh7 cells, while black indicates no change in gene expression. In the right panel, genes whose regulation showed a _P_ value of >0.01 are shown in gray.
FIG.4.
Increased activation of type I IFN receptor responses by REBOV infection. Panel A, immunofluorescence analysis of Huh7 cells infected with ZEBOV, REBOV, or MARV at an MOI of 0.1 for 24 h and then treated with 100 IU/ml of IFN-α-2b for an additional 24 h. Panel B, quantification of infectious viral particles in culture supernatants of infected cells in the presence and absence of 100 IU/ml IFN-α-2b (for 24 h) expressed as the log10 TCID50/ml. Panel C, scatter plots comparing the expression of genes that were >1.5× (P < 0.01) induced in mock-infected cells treated with 100 IU/ml IFN-α-2b for 24 h and in MARV-, ZEBOV-, and REBOV-infected cells (24 h) treated IFN-α-2b for 24 h. Expression of MX1 RNA is indicated by arrows. Panel D, matrix of genes induced >1.5× (P < 0.01) in ZEBOV-, REBOV-, and MARV-infected cells treated with IFN. Panel E, hierarchical clustering matrix of selected genes that were preferentially induced in REBOV-infected cells following IFN-α-2b treatment. In the top panel, genes shown in red were up-regulated and genes shown in green were down-regulated relative to uninfected Huh7 cells, and black indicates no change in gene expression. In the bottom panel, genes whose regulation showed a _P_ value of >0.01 are shown in gray. Panel F, Western blotting analysis showing expression of MX1 protein during ZEBOV, REBOV, and MARV infection in the absence and presence of IFN-α-2b.
FIG.4.
Increased activation of type I IFN receptor responses by REBOV infection. Panel A, immunofluorescence analysis of Huh7 cells infected with ZEBOV, REBOV, or MARV at an MOI of 0.1 for 24 h and then treated with 100 IU/ml of IFN-α-2b for an additional 24 h. Panel B, quantification of infectious viral particles in culture supernatants of infected cells in the presence and absence of 100 IU/ml IFN-α-2b (for 24 h) expressed as the log10 TCID50/ml. Panel C, scatter plots comparing the expression of genes that were >1.5× (P < 0.01) induced in mock-infected cells treated with 100 IU/ml IFN-α-2b for 24 h and in MARV-, ZEBOV-, and REBOV-infected cells (24 h) treated IFN-α-2b for 24 h. Expression of MX1 RNA is indicated by arrows. Panel D, matrix of genes induced >1.5× (P < 0.01) in ZEBOV-, REBOV-, and MARV-infected cells treated with IFN. Panel E, hierarchical clustering matrix of selected genes that were preferentially induced in REBOV-infected cells following IFN-α-2b treatment. In the top panel, genes shown in red were up-regulated and genes shown in green were down-regulated relative to uninfected Huh7 cells, and black indicates no change in gene expression. In the bottom panel, genes whose regulation showed a _P_ value of >0.01 are shown in gray. Panel F, Western blotting analysis showing expression of MX1 protein during ZEBOV, REBOV, and MARV infection in the absence and presence of IFN-α-2b.
FIG.4.
Increased activation of type I IFN receptor responses by REBOV infection. Panel A, immunofluorescence analysis of Huh7 cells infected with ZEBOV, REBOV, or MARV at an MOI of 0.1 for 24 h and then treated with 100 IU/ml of IFN-α-2b for an additional 24 h. Panel B, quantification of infectious viral particles in culture supernatants of infected cells in the presence and absence of 100 IU/ml IFN-α-2b (for 24 h) expressed as the log10 TCID50/ml. Panel C, scatter plots comparing the expression of genes that were >1.5× (P < 0.01) induced in mock-infected cells treated with 100 IU/ml IFN-α-2b for 24 h and in MARV-, ZEBOV-, and REBOV-infected cells (24 h) treated IFN-α-2b for 24 h. Expression of MX1 RNA is indicated by arrows. Panel D, matrix of genes induced >1.5× (P < 0.01) in ZEBOV-, REBOV-, and MARV-infected cells treated with IFN. Panel E, hierarchical clustering matrix of selected genes that were preferentially induced in REBOV-infected cells following IFN-α-2b treatment. In the top panel, genes shown in red were up-regulated and genes shown in green were down-regulated relative to uninfected Huh7 cells, and black indicates no change in gene expression. In the bottom panel, genes whose regulation showed a _P_ value of >0.01 are shown in gray. Panel F, Western blotting analysis showing expression of MX1 protein during ZEBOV, REBOV, and MARV infection in the absence and presence of IFN-α-2b.
FIG.4.
Increased activation of type I IFN receptor responses by REBOV infection. Panel A, immunofluorescence analysis of Huh7 cells infected with ZEBOV, REBOV, or MARV at an MOI of 0.1 for 24 h and then treated with 100 IU/ml of IFN-α-2b for an additional 24 h. Panel B, quantification of infectious viral particles in culture supernatants of infected cells in the presence and absence of 100 IU/ml IFN-α-2b (for 24 h) expressed as the log10 TCID50/ml. Panel C, scatter plots comparing the expression of genes that were >1.5× (P < 0.01) induced in mock-infected cells treated with 100 IU/ml IFN-α-2b for 24 h and in MARV-, ZEBOV-, and REBOV-infected cells (24 h) treated IFN-α-2b for 24 h. Expression of MX1 RNA is indicated by arrows. Panel D, matrix of genes induced >1.5× (P < 0.01) in ZEBOV-, REBOV-, and MARV-infected cells treated with IFN. Panel E, hierarchical clustering matrix of selected genes that were preferentially induced in REBOV-infected cells following IFN-α-2b treatment. In the top panel, genes shown in red were up-regulated and genes shown in green were down-regulated relative to uninfected Huh7 cells, and black indicates no change in gene expression. In the bottom panel, genes whose regulation showed a _P_ value of >0.01 are shown in gray. Panel F, Western blotting analysis showing expression of MX1 protein during ZEBOV, REBOV, and MARV infection in the absence and presence of IFN-α-2b.
FIG.4.
Increased activation of type I IFN receptor responses by REBOV infection. Panel A, immunofluorescence analysis of Huh7 cells infected with ZEBOV, REBOV, or MARV at an MOI of 0.1 for 24 h and then treated with 100 IU/ml of IFN-α-2b for an additional 24 h. Panel B, quantification of infectious viral particles in culture supernatants of infected cells in the presence and absence of 100 IU/ml IFN-α-2b (for 24 h) expressed as the log10 TCID50/ml. Panel C, scatter plots comparing the expression of genes that were >1.5× (P < 0.01) induced in mock-infected cells treated with 100 IU/ml IFN-α-2b for 24 h and in MARV-, ZEBOV-, and REBOV-infected cells (24 h) treated IFN-α-2b for 24 h. Expression of MX1 RNA is indicated by arrows. Panel D, matrix of genes induced >1.5× (P < 0.01) in ZEBOV-, REBOV-, and MARV-infected cells treated with IFN. Panel E, hierarchical clustering matrix of selected genes that were preferentially induced in REBOV-infected cells following IFN-α-2b treatment. In the top panel, genes shown in red were up-regulated and genes shown in green were down-regulated relative to uninfected Huh7 cells, and black indicates no change in gene expression. In the bottom panel, genes whose regulation showed a _P_ value of >0.01 are shown in gray. Panel F, Western blotting analysis showing expression of MX1 protein during ZEBOV, REBOV, and MARV infection in the absence and presence of IFN-α-2b.
FIG.4.
Increased activation of type I IFN receptor responses by REBOV infection. Panel A, immunofluorescence analysis of Huh7 cells infected with ZEBOV, REBOV, or MARV at an MOI of 0.1 for 24 h and then treated with 100 IU/ml of IFN-α-2b for an additional 24 h. Panel B, quantification of infectious viral particles in culture supernatants of infected cells in the presence and absence of 100 IU/ml IFN-α-2b (for 24 h) expressed as the log10 TCID50/ml. Panel C, scatter plots comparing the expression of genes that were >1.5× (P < 0.01) induced in mock-infected cells treated with 100 IU/ml IFN-α-2b for 24 h and in MARV-, ZEBOV-, and REBOV-infected cells (24 h) treated IFN-α-2b for 24 h. Expression of MX1 RNA is indicated by arrows. Panel D, matrix of genes induced >1.5× (P < 0.01) in ZEBOV-, REBOV-, and MARV-infected cells treated with IFN. Panel E, hierarchical clustering matrix of selected genes that were preferentially induced in REBOV-infected cells following IFN-α-2b treatment. In the top panel, genes shown in red were up-regulated and genes shown in green were down-regulated relative to uninfected Huh7 cells, and black indicates no change in gene expression. In the bottom panel, genes whose regulation showed a _P_ value of >0.01 are shown in gray. Panel F, Western blotting analysis showing expression of MX1 protein during ZEBOV, REBOV, and MARV infection in the absence and presence of IFN-α-2b.
FIG.5.
Phosphorylation of STAT proteins in filovirus-infected cells following type I IFN treatment. Western blotting analysis was performed on equal-mass cell lysates obtained from mock-, SeV-, ZEBOV-, REBOV-, or MARV-infected cells at 24 h in the absence or presence of 100 IU/ml IFN-α-2b for 30 min, using antibodies specific for phosphorylated STAT1 or total STAT1 protein (A) or for phosphorylated STAT2 or total STAT2 protein (B).
Similar articles
- Lethality and pathogenesis of airborne infection with filoviruses in A129 α/β -/- interferon receptor-deficient mice.
Lever MS, Piercy TJ, Steward JA, Eastaugh L, Smither SJ, Taylor C, Salguero FJ, Phillpotts RJ. Lever MS, et al. J Med Microbiol. 2012 Jan;61(Pt 1):8-15. doi: 10.1099/jmm.0.036210-0. Epub 2011 Aug 18. J Med Microbiol. 2012. PMID: 21852521 - Filovirus infection of STAT-1 knockout mice.
Raymond J, Bradfute S, Bray M. Raymond J, et al. J Infect Dis. 2011 Nov;204 Suppl 3:S986-90. doi: 10.1093/infdis/jir335. J Infect Dis. 2011. PMID: 21987780 - Effects of Filovirus Interferon Antagonists on Responses of Human Monocyte-Derived Dendritic Cells to RNA Virus Infection.
Yen BC, Basler CF. Yen BC, et al. J Virol. 2016 Apr 29;90(10):5108-5118. doi: 10.1128/JVI.00191-16. Print 2016 May 15. J Virol. 2016. PMID: 26962215 Free PMC article. - Current knowledge on lower virulence of Reston Ebola virus (in French: Connaissances actuelles sur la moindre virulence du virus Ebola Reston).
Morikawa S, Saijo M, Kurane I. Morikawa S, et al. Comp Immunol Microbiol Infect Dis. 2007 Sep;30(5-6):391-8. doi: 10.1016/j.cimid.2007.05.005. Epub 2007 Jul 3. Comp Immunol Microbiol Infect Dis. 2007. PMID: 17610952 Review. - Filoviral immune evasion mechanisms.
Ramanan P, Shabman RS, Brown CS, Amarasinghe GK, Basler CF, Leung DW. Ramanan P, et al. Viruses. 2011 Sep;3(9):1634-49. doi: 10.3390/v3091634. Epub 2011 Sep 7. Viruses. 2011. PMID: 21994800 Free PMC article. Review.
Cited by
- Altered microRNA Transcriptome in Cultured Human Liver Cells upon Infection with Ebola Virus.
Diallo I, Ho J, Laffont B, Laugier J, Benmoussa A, Lambert M, Husseini Z, Soule G, Kozak R, Kobinger GP, Provost P. Diallo I, et al. Int J Mol Sci. 2021 Apr 6;22(7):3792. doi: 10.3390/ijms22073792. Int J Mol Sci. 2021. PMID: 33917562 Free PMC article. - Differential transcriptional responses to Ebola and Marburg virus infection in bat and human cells.
Hölzer M, Krähling V, Amman F, Barth E, Bernhart SH, Carmelo VA, Collatz M, Doose G, Eggenhofer F, Ewald J, Fallmann J, Feldhahn LM, Fricke M, Gebauer J, Gruber AJ, Hufsky F, Indrischek H, Kanton S, Linde J, Mostajo N, Ochsenreiter R, Riege K, Rivarola-Duarte L, Sahyoun AH, Saunders SJ, Seemann SE, Tanzer A, Vogel B, Wehner S, Wolfinger MT, Backofen R, Gorodkin J, Grosse I, Hofacker I, Hoffmann S, Kaleta C, Stadler PF, Becker S, Marz M. Hölzer M, et al. Sci Rep. 2016 Oct 7;6:34589. doi: 10.1038/srep34589. Sci Rep. 2016. PMID: 27713552 Free PMC article. - The VP35 protein of Ebola virus inhibits the antiviral effect mediated by double-stranded RNA-dependent protein kinase PKR.
Feng Z, Cerveny M, Yan Z, He B. Feng Z, et al. J Virol. 2007 Jan;81(1):182-92. doi: 10.1128/JVI.01006-06. Epub 2006 Oct 25. J Virol. 2007. PMID: 17065211 Free PMC article. - Processing of genome 5' termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction.
Habjan M, Andersson I, Klingström J, Schümann M, Martin A, Zimmermann P, Wagner V, Pichlmair A, Schneider U, Mühlberger E, Mirazimi A, Weber F. Habjan M, et al. PLoS One. 2008 Apr 30;3(4):e2032. doi: 10.1371/journal.pone.0002032. PLoS One. 2008. PMID: 18446221 Free PMC article. - Functional genomics reveals the induction of inflammatory response and metalloproteinase gene expression during lethal Ebola virus infection.
Cilloniz C, Ebihara H, Ni C, Neumann G, Korth MJ, Kelly SM, Kawaoka Y, Feldmann H, Katze MG. Cilloniz C, et al. J Virol. 2011 Sep;85(17):9060-8. doi: 10.1128/JVI.00659-11. Epub 2011 Jul 6. J Virol. 2011. PMID: 21734050 Free PMC article.
References
- Baize, S., E. M. Leroy, M. C. Georges-Courbot, M. Capron, J. Lansoud-Soukate, P. Debre, S. P. Fisher-Hoch, J. B. McCormick, and A. J. Georges. 1999. Defective humoral responses and extensive intravascular apoptosis are associated with fatal outcome in Ebola virus-infected patients. Nat. Med. 5:423-426. - PubMed
- Baize, S., E. M. Leroy, E. Mavoungou, and S. P. Fisher-Hoch. 2000. Apoptosis in fatal Ebola infection. Does the virus toll the bell for immune system? Apoptosis 5:5-7. - PubMed
- Basler, C. F., and A. Garcia-Sastre. 2002. Viruses and the type I interferon antiviral system: induction and evasion. Int. Rev. Immunol. 21:305-337. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI056214-02/AI/NIAID NIH HHS/United States
- AI058113-01/AI/NIAID NIH HHS/United States
- P30 DA015625/DA/NIDA NIH HHS/United States
- DA15625-03/DA/NIDA NIH HHS/United States
- R21 AI056214/AI/NIAID NIH HHS/United States
- P01 AI058113/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous