Nidovirales: evolving the largest RNA virus genome - PubMed (original) (raw)
Review
Nidovirales: evolving the largest RNA virus genome
Alexander E Gorbalenya et al. Virus Res. 2006 Apr.
Abstract
This review focuses on the monophyletic group of animal RNA viruses united in the order Nidovirales. The order includes the distantly related coronaviruses, toroviruses, and roniviruses, which possess the largest known RNA genomes (from 26 to 32kb) and will therefore be called "large" nidoviruses in this review. They are compared with their arterivirus cousins, which also belong to the Nidovirales despite having a much smaller genome (13-16kb). Common and unique features that have been identified for either large or all nidoviruses are outlined. These include the nidovirus genetic plan and genome diversity, the composition of the replicase machinery and virus particles, virus-specific accessory genes, the mechanisms of RNA and protein synthesis, and the origin and evolution of nidoviruses with small and large genomes. Nidoviruses employ single-stranded, polycistronic RNA genomes of positive polarity that direct the synthesis of the subunits of the replicative complex, including the RNA-dependent RNA polymerase and helicase. Replicase gene expression is under the principal control of a ribosomal frameshifting signal and a chymotrypsin-like protease, which is assisted by one or more papain-like proteases. A nested set of subgenomic RNAs is synthesized to express the 3'-proximal ORFs that encode most conserved structural proteins and, in some large nidoviruses, also diverse accessory proteins that may promote virus adaptation to specific hosts. The replicase machinery includes a set of RNA-processing enzymes some of which are unique for either all or large nidoviruses. The acquisition of these enzymes may have improved the low fidelity of RNA replication to allow genome expansion and give rise to the ancestors of small and, subsequently, large nidoviruses.
Figures
Fig. 1
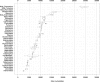
Distribution of genome sizes of ssRNA+ viruses. Box-and-whisker graphs were used to plot the family/group-specific distribution of genome sizes of all ssRNA+ viruses whose genome sequences have been placed in the NCBI Viral Genome Resource (Bao et al., 2004) by December 7, 2005 (Faase and Gorbalenya, unpublished data). Four major groups of nidoviruses are highlighted with the Nido- prefix, and the Coronaviridae family is split into the Nido-Coronavirus and the Nido-Torovirus groups. The box spans from the first to the third quartile and includes the median, indicated by the vertical line. The whiskers extend to the extreme values that are distant from the box at most 1.5 times the interquartile range. Values beyond this distance are indicated by circles (outliers).
Fig. 2
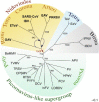
RdRp-based RNA virus tree that includes nidoviruses. The most conserved part of RdRps from representative viruses in the Picornaviridae, Dicistroviridae, Sequiviridae, Comoviridae, Caliciviridae, Potyviridae, Coronaviridae, Roniviridae, Arteriviridae, Birnaviridae, Tetraviridae and unclassified insect viruses was aligned. An unrooted neighbour-joining tree was inferred using the ClustalX1.81 software. For details of the analysis, see Gorbalenya et al. (2002). All bifurcations with support in >700 out of 1000 bootstraps are indicated. Different groups of viruses are highlighted with different colours. The tree was modified from (Gorbalenya et al., 2002, Snijder et al., 2005b, Spaan et al., 2005b). Virus families/groups and abbreviations of viruses included in the analysis are as follows: Coronaviridae: avian infectious bronchitis virus (IBV), severe acute respiratory syndrome virus (SARS-CoV) and Equine torovirus (EToV); Arteriviridae, Equine arteritis virus (EAV) and Porcine reproductive and respiratory syndrome virus strain VR-2332 (PRRSV); Roniviridae: Gill-associated virus (GAV); Picornaviridae, human poliovirus type 3 Leon strain (PV) and parechovirus 1 (HPeV); Iflavirus, infectious flacherie virus (InFV); unclassified insect viruses, Acyrthosiphon pisum virus (APV); Dicistroviridae, Drosophila C virus (DCV); Sequiviridae, Rice tungro spherical virus (RTSV) and Parsnip yellow fleck virus (PYFV); Comoviridae, Cowpea severe mosaic virus (CPSMV) and Tobacco ringspot virus (TRSV); Caliciviridae, Feline calicivirus F9 (FCV) and Lordsdale virus (LORDV); Potyviridae, Tobacco vein mottling virus (TVMV) and Barley mild mosaic virus (BaMMV); Tetraviridae, Thosea asigna virus (TaV) and Euprosterna elaeasa virus (EeV); Birnaviridae, infectious pancreatic necrosis virus (IPNV) and infectious bursal disease virus (IBDV). A plausible direction to the root of the nidovirus domain is indicated (red arrow). As discussed in the text, arteriviruses rather than roniviruses might have been the first to branch off from the nidovirus trunk. This revised topology of the two lineages is also indicated (blue arrows).
Fig. 3
Genome organization of selected nidoviruses. The genomic ORFs of viruses representing major lineages of nidoviruses are indicated and the names of the replicase and main virion genes are given. References to the nomenclature of accessory genes can be found in the text. Genomes of large and small nidoviruses are drawn to different scales. This figure was updated from Fig. 39.2 presented in Siddell et al. (2005).
Fig. 4
Domain organization of the replicase pp1ab polyprotein for selected nidoviruses. Shown are currently mapped domains in the pp1ab replicase polyproteins of viruses representing major lineages of nidoviruses. Arrows represent sites in pp1ab that are cleaved by papain-like proteinases (orange and blue) or chymotrypsin-like (3CLpro) proteinase (red). For viruses with the full cleavage site map available (arteriviruses and coronaviruses), the proteolytic cleavage products are numbered. A tentative cleavage site map for EToV is from an unpublished work by A.E. Gorbalenya. Within the cleavage products, the location and names of domains that have been identified as structurally and functionally related are highlighted. These include diverse domains with conserved Cys and His residues (C/H), putative transmembrane domains (TM), domains with conserved features (AC, X and Y), and domains that have been associated with proteolysis (PL1, PL2, and 3CL), RNA-dependent RNA synthesis (RdRp), helicase (HEL), exonuclease (ExoN), uridylate-specific endoribonuclease (N; NendoU in the main text), methyl transferase (MT) and cyclic phosphodiesterase (CPD) activities. Note that due to space limitations, the domain names in this figure may be abbreviated derivatives from those used in the main text. Polyproteins of large and small nidoviruses are drawn to different scales. This figure was updated from Fig. 39.4 presented in Siddell et al. (2005).
Fig. 5
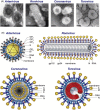
Architecture of particles of members of the order Nidovirales: electron micrographs (A) and schematic representations (B). N, nucleocapsid protein; S, spike protein; M, membrane protein; E, envelope protein; HE, hemagglutinin-esterase. Coronavirus M protein interacts with the N protein. In arterivirus, GP5 and M are major envelope proteins, while GP2, GP3, GP4, and E are minor envelope proteins. Toro- and roniviruses lack the E protein present in corona- and arteriviruses. An equivalent of the M protein has not yet been identified in roniviruses (although see also Fig. 3). This figure was reproduced from Fig. 20.1 presented in Snijder et al. (2005b). Images were reproduced (with permission) from (Snijder and Muelenberg, 1998) (arterivirus), (Spann et al., 1995) (ronivirus), Dr. Fred Murphy, Centers for Disease Control and Prevention (CDC), Atlanta, USA (coronavirus) and (Weiss et al., 1983) (torovirus).
Fig. 6
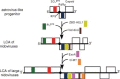
Origin and evolution of the nidovirus genome plan. A tentative evolutionary scenario leading to the origin of the LCAs of nidoviruses and large nidoviruses from a progenitor with an astrovirus-like genome organization is illustrated. The three shown genomes are fictitious, although they are drawn to a relative common size scale and include major replicative domains found in genomes of respective contemporary viruses, as discussed in the text.
Fig. 7
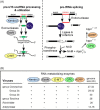
RNA-processing enzymes of nidoviruses: possible cooperation and virus distribution. (A) Cellular pathways for processing of pre-U16 small nucleolar RNA (snoRNA) and pre-tRNA splicing in which homologs of five nidovirus RNA-processing enzymes are involved. Note that both pathways produce intermediates with 2′-3′-cyclic phosphate termini (blue circles), indicating the structural basis for a possible cooperation of the nidovirus homologs in a single pathway (Snijder et al., 2003). (B) Table summarizing the conservation of five (putative) RNA-processing enzymes among representatives of large and small nidoviruses. This figure was updated from Fig. 5 presented in Snijder et al. (2003).
Similar articles
- Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes.
Nga PT, Parquet Mdel C, Lauber C, Parida M, Nabeshima T, Yu F, Thuy NT, Inoue S, Ito T, Okamoto K, Ichinose A, Snijder EJ, Morita K, Gorbalenya AE. Nga PT, et al. PLoS Pathog. 2011 Sep;7(9):e1002215. doi: 10.1371/journal.ppat.1002215. Epub 2011 Sep 8. PLoS Pathog. 2011. PMID: 21931546 Free PMC article. - Characterization of White bream virus reveals a novel genetic cluster of nidoviruses.
Schütze H, Ulferts R, Schelle B, Bayer S, Granzow H, Hoffmann B, Mettenleiter TC, Ziebuhr J. Schütze H, et al. J Virol. 2006 Dec;80(23):11598-609. doi: 10.1128/JVI.01758-06. Epub 2006 Sep 20. J Virol. 2006. PMID: 16987966 Free PMC article. - Nidovirus RNA polymerases: Complex enzymes handling exceptional RNA genomes.
Posthuma CC, Te Velthuis AJW, Snijder EJ. Posthuma CC, et al. Virus Res. 2017 Apr 15;234:58-73. doi: 10.1016/j.virusres.2017.01.023. Epub 2017 Feb 6. Virus Res. 2017. PMID: 28174054 Free PMC article. Review. - A planarian nidovirus expands the limits of RNA genome size.
Saberi A, Gulyaeva AA, Brubacher JL, Newmark PA, Gorbalenya AE. Saberi A, et al. PLoS Pathog. 2018 Nov 1;14(11):e1007314. doi: 10.1371/journal.ppat.1007314. eCollection 2018 Nov. PLoS Pathog. 2018. PMID: 30383829 Free PMC article. - Structure and Expression of Large (+)RNA Genomes of Viruses of Higher Eukaryotes.
Agranovsky AA. Agranovsky AA. Biochemistry (Mosc). 2021 Mar;86(3):248-261. doi: 10.1134/S0006297921030020. Biochemistry (Mosc). 2021. PMID: 33838627 Free PMC article. Review.
Cited by
- Bioinformatics and functional analyses of coronavirus nonstructural proteins involved in the formation of replicative organelles.
Neuman BW. Neuman BW. Antiviral Res. 2016 Nov;135:97-107. doi: 10.1016/j.antiviral.2016.10.005. Epub 2016 Oct 13. Antiviral Res. 2016. PMID: 27743916 Free PMC article. Review. - Physiologic RNA targets and refined sequence specificity of coronavirus EndoU.
Ancar R, Li Y, Kindler E, Cooper DA, Ransom M, Thiel V, Weiss SR, Hesselberth JR, Barton DJ. Ancar R, et al. RNA. 2020 Dec;26(12):1976-1999. doi: 10.1261/rna.076604.120. Epub 2020 Sep 28. RNA. 2020. PMID: 32989044 Free PMC article. - Novel RT-ddPCR assays for simultaneous quantification of multiple noncoding and coding regions of SARS-CoV-2 RNA.
Telwatte S, Kumar N, Vallejo-Gracia A, Kumar GR, Lu CM, Ott M, Wong JK, Yukl SA. Telwatte S, et al. J Virol Methods. 2021 Jun;292:114115. doi: 10.1016/j.jviromet.2021.114115. Epub 2021 Mar 2. J Virol Methods. 2021. PMID: 33667568 Free PMC article. - Design of Potential RNAi (miRNA and siRNA) Molecules for Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Gene Silencing by Computational Method.
Nur SM, Hasan MA, Amin MA, Hossain M, Sharmin T. Nur SM, et al. Interdiscip Sci. 2015 Sep;7(3):257-65. doi: 10.1007/s12539-015-0266-9. Epub 2015 Jul 30. Interdiscip Sci. 2015. PMID: 26223545 Free PMC article. - Hidden genomic diversity of SARS-CoV-2: implications for qRT-PCR diagnostics and transmission.
Sapoval N, Mahmoud M, Jochum MD, Liu Y, Elworth RAL, Wang Q, Albin D, Ogilvie H, Lee MD, Villapol S, Hernandez KM, Berry IM, Foox J, Beheshti A, Ternus K, Aagaard KM, Posada D, Mason CE, Sedlazeck F, Treangen TJ. Sapoval N, et al. bioRxiv [Preprint]. 2020 Jul 2:2020.07.02.184481. doi: 10.1101/2020.07.02.184481. bioRxiv. 2020. PMID: 32637955 Free PMC article. Updated. Preprint.
References
- Allen M.D., Buckle A.M., Cordell S.C., Lowe J., Bycroft M. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of MacroH2A. J. Mol. Biol. 2003;330:503–511. - PubMed
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources