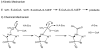Functional annotation and kinetic characterization of PhnO from Salmonella enterica - PubMed (original) (raw)
Functional annotation and kinetic characterization of PhnO from Salmonella enterica
James C Errey et al. Biochemistry. 2006.
Abstract
Phosphorus is an essential nutrient for all living organisms. Under conditions of inorganic phosphate starvation, genes from the Pho regulon are induced, allowing microorganisms to use phosphonates as a source of phosphorus. The phnO gene was previously annotated as a transcriptional regulator of unknown function due to sequence homology with members of the GCN5-related N-acyltransferase family (GNAT). PhnO can now be functionally annotated as an aminoalkylphosphonic acid N-acetyltransferase which is able to acetylate a range of aminoalkylphosphonic acids. Studies revealed that PhnO proceeds via an ordered, sequential kinetic mechanism with AcCoA binding first followed by aminoalkylphosphonate. Attack by the amine on the thioester of AcCoA generates the tetrahedral intermediate that collapses to generate the products. The enzyme also requires a divalent metal ion for activity, which is the first example of this requirement for a GNAT family member.
Figures
Figure 1
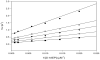
Double reciprocal plot of initial rate data at varying _S_-1AEP concentrations and fixed concentrations of acetyl-CoA at 10 μM (♀), 20 μM (○), 30 μM ( ), 60 μM (→) and 250 μM (†). The pattern of intersecting lines is indicative of a sequential kinetic mechanism.
), 60 μM (→) and 250 μM (†). The pattern of intersecting lines is indicative of a sequential kinetic mechanism.
Figure 2
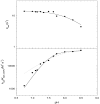
Dependence of _k_cat and _k_cat/K _S_-1AEP on pH at saturating levels of acetyl-CoA and Ni2+. The dotted and solid lines are fits to Eqs 7 and 8, respectively. The _k_cat pH profile revealed the presence a single ionizable group with a pK value of 8.2 (± 0.1). The _k_cat/K _S_-1AEP profile was fit to Eq. 8, which describes the dependence on 2 groups with similar pK values of 7.5 (± 0.1) and Eq. 7, which describes the dependence on 1 group (dotted line).
Figure 3
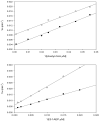
Reciprocal plots of the solvent kinetic isotope effect determined for PhnO using acetyl-CoA (top panel) or _S_-1AEP (bottom panel) as the variable substrate, in the presence of saturating concentrations of Ni2+. The symbols are the experimentally determined values in H2O (♀)or 90% D2O ( ), while the lines are fits of the data to Eq. 9.)
), while the lines are fits of the data to Eq. 9.)
Scheme 1
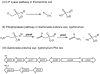
Pathways for phosphonate catabolism. (I) The C-P lyase pathway acts on alkylphosphonates as well as aminoalkylphosphonates. (II) The phosphonatase pathway that uses 2-aminoethylphosphonate and cleaves the C-P bond in a two-step transamination and hydrolysis process. (III) The S. enterica phn gene locus; phnR encodes a putative aminoalkylphosphonate transport repressor, phnS-V encode putative aminoalkylphosphonate transport components, phnW encodes a 2-aminoethylphosphonate transaminase, phnX encodes a 2-phosphonoacetaldehyde hydrolase, phnA encodes a putative phosphonoacetate hydrolase, phnB encodes a putative phosphonoacetate transporter, phnO encodes an aminoalkylphosphonic _N_-acetyltransferase.
Scheme 2
Proposed kinetic and chemical mechanisms for aminophosphonate _N_-acetylation by PhnO
Similar articles
- Kinetic mechanism of human histone acetyltransferase P/CAF.
Tanner KG, Langer MR, Denu JM. Tanner KG, et al. Biochemistry. 2000 Oct 3;39(39):11961-9. doi: 10.1021/bi001272h. Biochemistry. 2000. PMID: 11009610 - Kinetic and mutagenic characterization of the chromosomally encoded Salmonella enterica AAC(6')-Iy aminoglycoside N-acetyltransferase.
Magnet S, Lambert T, Courvalin P, Blanchard JS. Magnet S, et al. Biochemistry. 2001 Mar 27;40(12):3700-9. doi: 10.1021/bi002736e. Biochemistry. 2001. PMID: 11297438 - Classification and phylogeny for the annotation of novel eukaryotic GNAT acetyltransferases.
Krtenic B, Drazic A, Arnesen T, Reuter N. Krtenic B, et al. PLoS Comput Biol. 2020 Dec 23;16(12):e1007988. doi: 10.1371/journal.pcbi.1007988. eCollection 2020 Dec. PLoS Comput Biol. 2020. PMID: 33362253 Free PMC article. - The serine acetyltransferase reaction: acetyl transfer from an acylpantothenyl donor to an alcohol.
Johnson CM, Roderick SL, Cook PF. Johnson CM, et al. Arch Biochem Biophys. 2005 Jan 1;433(1):85-95. doi: 10.1016/j.abb.2004.08.014. Arch Biochem Biophys. 2005. PMID: 15581568 Review. - Structure and functions of the GNAT superfamily of acetyltransferases.
Vetting MW, S de Carvalho LP, Yu M, Hegde SS, Magnet S, Roderick SL, Blanchard JS. Vetting MW, et al. Arch Biochem Biophys. 2005 Jan 1;433(1):212-26. doi: 10.1016/j.abb.2004.09.003. Arch Biochem Biophys. 2005. PMID: 15581578 Review.
Cited by
- Phosphonate biosynthesis and catabolism: a treasure trove of unusual enzymology.
Peck SC, van der Donk WA. Peck SC, et al. Curr Opin Chem Biol. 2013 Aug;17(4):580-8. doi: 10.1016/j.cbpa.2013.06.018. Epub 2013 Jul 17. Curr Opin Chem Biol. 2013. PMID: 23870698 Free PMC article. Review. - Metatranscriptomic and functional metagenomic analysis of methylphosphonate utilization by marine bacteria.
Martínez A, Ventouras LA, Wilson ST, Karl DM, Delong EF. Martínez A, et al. Front Microbiol. 2013 Nov 26;4:340. doi: 10.3389/fmicb.2013.00340. eCollection 2013. Front Microbiol. 2013. PMID: 24324460 Free PMC article. - Bacterial GCN5-Related N-Acetyltransferases: From Resistance to Regulation.
Favrot L, Blanchard JS, Vergnolle O. Favrot L, et al. Biochemistry. 2016 Feb 23;55(7):989-1002. doi: 10.1021/acs.biochem.5b01269. Epub 2016 Feb 9. Biochemistry. 2016. PMID: 26818562 Free PMC article. Review. - Catabolism and detoxification of 1-aminoalkylphosphonic acids: N-acetylation by the phnO gene product.
Hove-Jensen B, McSorley FR, Zechel DL. Hove-Jensen B, et al. PLoS One. 2012;7(10):e46416. doi: 10.1371/journal.pone.0046416. Epub 2012 Oct 3. PLoS One. 2012. PMID: 23056305 Free PMC article. - Pulse-chase studies of the synthesis of acetyl-CoA by carbon monoxide dehydrogenase/acetyl-CoA synthase: evidence for a random mechanism of methyl and carbonyl addition.
Seravalli J, Ragsdale SW. Seravalli J, et al. J Biol Chem. 2008 Mar 28;283(13):8384-94. doi: 10.1074/jbc.M709470200. Epub 2008 Jan 18. J Biol Chem. 2008. PMID: 18203715 Free PMC article.
References
- Vershinina OA, Znamenskaia LV. The Pho regulons of bacteria. Mikrobiologiia. 2002;71:581–95. - PubMed
- Kononova SV, Nesmeyanova MA. Phosphonates and their degradation by microorganisms. Biochemistry (Mosc) 2002;67:184–95. - PubMed
- Kennedy KE, Thompson GA., Jr. Phosphonolipids: localization in surface membranes of Tetrahymena. Science. 1970;168:989–91. - PubMed
- Wanner BL. Molecular genetics of carbon-phosphorus bond cleavage in bacteria. Biodegradation. 1994;5:175–84. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources