Solution structure of the severe acute respiratory syndrome-coronavirus heptad repeat 2 domain in the prefusion state - PubMed (original) (raw)
Solution structure of the severe acute respiratory syndrome-coronavirus heptad repeat 2 domain in the prefusion state
Susanna Hakansson-McReynolds et al. J Biol Chem. 2006.
Abstract
The envelope glycoprotein, termed the spike protein, of severe acute respiratory syndrome coronavirus (SARS-CoV) is known to mediate viral entry. Similar to other class 1 viral fusion proteins, the heptad repeat regions of SARS-CoV spike are thought to undergo conformational changes from a prefusion form to a subsequent post-fusion form that enables fusion of the viral and host membranes. Recently, the structure of a post-fusion form of SARS-CoV spike, which consists of isolated domains of heptad repeats 1 and 2 (HR1 and HR2), has been determined by x-ray crystallography. To date there is no structural information for the prefusion conformations of SARS-CoV HR1 and HR2. In this work we present the NMR structure of the HR2 domain (residues 1141-1193) from SARS-CoV (termed S2-HR2) in the presence of the co-solvent trifluoroethanol. We find that in the absence of HR1, S2-HR2 forms a coiled coil symmetric trimer with a complex molecular mass of 18 kDa. The S2-HR2 structure, which is the first example of the prefusion form of coronavirus envelope, supports the current model of viral membrane fusion and gives insight into the design of structure-based antagonists of SARS.
Figures
FIGURE 1
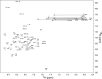
15N-edited HSQC of SARS-CoV S2-HR2. The sample conditions were 1 m
m
S2-HR2 monomer, 10 m
m
NaPO4, pH 7.0, 30% TFE-d3 at 25 °C. Horizontal lines connect the side chain amide proton pairs of asparagine and glutamine residues.
FIGURE 2
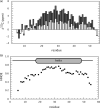
Secondary structure of SARS-CoV S2-HR2.a, secondary chemical shift of S2-HR2 13Cα (gray) and 13C′ (white) with respect to random coil values. Random coil values were taken from Wishart and Case (64). b, HNOE of S2-HR2.
FIGURE 3
CLEANEX-PM 15N-edited HSQC. The sample conditions were 1 m
m
of S2-HR2 monomer, 10 m
m
NaPO4, pH 7.0, 30% TFE-d3 at 25 °C with a mixing time of 100 ms. Note that residues Thr4 and Ser5, which occur in an unstructured region at the N terminus, exhibit negative contours, presumably because of ROE effects and/or intermolecular NOEs with H2O (49). The absence of a correlations for residues 17-47 suggests that they are involved in H-bonds. The absence of correlations for residues 14-16 and 50 may suggest the presence of H-bonds. The other missing correlations include those of residues 1-3, which are also missing from the 15N-edited HSQC (residue 6 is a proline).
FIGURE 4
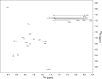
Intermolecular NOEs of SARS-CoV S2-HR2 showing intersubunit contacts involving the helix. Selected strips from the three-dimensional F1-filtered F2-edited 1H-13C NOESY-HSQC spectrum (mixing time of 120 ms) recorded on a 1:1 mixture of 12C/14N-and 13C/15N-labled S2-HR2.
FIGURE 5
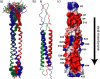
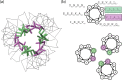
Solution structure of SARS-CoV S2-HR2.a, ensemble of 30 low energy structures of showing the superimposition of the backbone atoms. b, ribbon representation of the minimized mean structure. c, electrostatic map of the minimized mean structure. In a and b, subunits A, B, and C are colored red, green, and blue, respectively. The direction of the viral membrane is shown by an arrow.
FIGURE 6
Structural features of SARS-CoV S2-HR2.a, intermolecular contacts between the S2-HR helices. b, helical wheel representation of S2-HR2 residues Asn17-Leu47 looking down the helical axis, starting at the N-terminal end. There are seven residues/heptad where the individual positions of the seven residues are denoted by the letters a-f. The residues in the a and d positions make up the hydrophobic interface of the trimeric coiled coil of the S2-HR2 structure. The a positions are highlighted purple, and the d positions are highlighted green.
Similar articles
- Characterization of the heptad repeat regions, HR1 and HR2, and design of a fusion core structure model of the spike protein from severe acute respiratory syndrome (SARS) coronavirus.
Xu Y, Zhu J, Liu Y, Lou Z, Yuan F, Liu Y, Cole DK, Ni L, Su N, Qin L, Li X, Bai Z, Bell JI, Pang H, Tien P, Gao GF, Rao Z. Xu Y, et al. Biochemistry. 2004 Nov 9;43(44):14064-71. doi: 10.1021/bi049101q. Biochemistry. 2004. PMID: 15518555 - Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors.
Liu S, Xiao G, Chen Y, He Y, Niu J, Escalante CR, Xiong H, Farmar J, Debnath AK, Tien P, Jiang S. Liu S, et al. Lancet. 2004 Mar 20;363(9413):938-47. doi: 10.1016/S0140-6736(04)15788-7. Lancet. 2004. PMID: 15043961 Free PMC article. - Characterization of the prefusion and transition states of severe acute respiratory syndrome coronavirus S2-HR2.
McReynolds S, Jiang S, Guo Y, Celigoy J, Schar C, Rong L, Caffrey M. McReynolds S, et al. Biochemistry. 2008 Jul 1;47(26):6802-8. doi: 10.1021/bi800622t. Epub 2008 Jun 10. Biochemistry. 2008. PMID: 18540634 - Severe acute respiratory syndrome coronavirus entry into host cells: Opportunities for therapeutic intervention.
Yeung KS, Yamanaka GA, Meanwell NA. Yeung KS, et al. Med Res Rev. 2006 Jul;26(4):414-33. doi: 10.1002/med.20055. Med Res Rev. 2006. PMID: 16521129 Free PMC article. Review. - Multimerization of the heptad repeat regions of the SARS-CoV 2 spike protein.
Aisenbrey C, Bechinger B. Aisenbrey C, et al. Biochim Biophys Acta Biomembr. 2024 Feb;1866(2):184259. doi: 10.1016/j.bbamem.2023.184259. Epub 2023 Dec 5. Biochim Biophys Acta Biomembr. 2024. PMID: 38061554 Review.
Cited by
- Identification of a new region of SARS-CoV S protein critical for viral entry.
Guo Y, Tisoncik J, McReynolds S, Farzan M, Prabhakar BS, Gallagher T, Rong L, Caffrey M. Guo Y, et al. J Mol Biol. 2009 Dec 11;394(4):600-5. doi: 10.1016/j.jmb.2009.10.032. Epub 2009 Oct 21. J Mol Biol. 2009. PMID: 19853613 Free PMC article. - Biophysical characterization of HRC peptide analogs interaction with heptad repeat regions of the SARS-coronavirus Spike fusion protein core.
Yan Z, Tripet B, Hodges RS. Yan Z, et al. J Struct Biol. 2006 Aug;155(2):162-75. doi: 10.1016/j.jsb.2006.03.024. Epub 2006 Apr 27. J Struct Biol. 2006. PMID: 16765058 Free PMC article. - The human coronavirus HCoV-229E S-protein structure and receptor binding.
Li Z, Tomlinson AC, Wong AH, Zhou D, Desforges M, Talbot PJ, Benlekbir S, Rubinstein JL, Rini JM. Li Z, et al. Elife. 2019 Oct 25;8:e51230. doi: 10.7554/eLife.51230. Elife. 2019. PMID: 31650956 Free PMC article. - Allosteric perspective on the mutability and druggability of the SARS-CoV-2 Spike protein.
Tan ZW, Tee WV, Samsudin F, Guarnera E, Bond PJ, Berezovsky IN. Tan ZW, et al. Structure. 2022 Apr 7;30(4):590-607.e4. doi: 10.1016/j.str.2021.12.011. Epub 2022 Jan 20. Structure. 2022. PMID: 35063064 Free PMC article. - HIV envelope: challenges and opportunities for development of entry inhibitors.
Caffrey M. Caffrey M. Trends Microbiol. 2011 Apr;19(4):191-7. doi: 10.1016/j.tim.2011.02.001. Epub 2011 Mar 4. Trends Microbiol. 2011. PMID: 21377881 Free PMC article. Review.
References
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. N. Engl. J. Med. 2003;348:1953–1966. - PubMed
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. Science. 2003;300:1399–1404. - PubMed
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Science. 2003;300:1394–1399. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous