Distinct mechanisms control the stability of the related S-phase cyclins Clb5 and Clb6 - PubMed (original) (raw)
Distinct mechanisms control the stability of the related S-phase cyclins Clb5 and Clb6
Leisa P Jackson et al. Mol Cell Biol. 2006 Mar.
Abstract
The yeast S-phase cyclins Clb5 and Clb6 are closely related proteins that are synthesized late in G1. Although often grouped together with respect to function, Clb5 and Clb6 exhibit differences in their ability to promote S-phase progression. DNA replication is significantly slowed in clb5Delta mutants but not in clb6Delta mutants. We have examined the basis for the differential functions of Clb5 and Clb6 and determined that unlike Clb5, which is stable until mitosis, Clb6 is degraded rapidly at the G1/S border. N-terminal deletions of CLB6 were hyperstabilized, suggesting that the sequences responsible for directing the destruction of Clb6 reside in the N terminus. Clb6 lacks the destruction box motif responsible for the anaphase promoting complex-mediated destruction of Clb5 but contains putative Cdc4 degron motifs in the N terminus. Clb6 was hyperstabilized in cdc34-3 and cdc4-3 mutants at restrictive temperatures and when S/T-P phosphorylation sites in the N terminus were mutated to nonphosphorylatable residues. Efficient degradation of Clb6 requires the activities of both Cdc28 and Pho85. Finally, hyperstabilized Clb6 expressed from the CLB6 promoter rescued the slow S-phase defect exhibited by clb5Delta cells. Taken together, these findings suggest that the SCF(Cdc4) ubiquitin ligase complex regulates Clb6 turnover and that the functional differences exhibited by Clb5 and Clb6 arise from the distinct mechanisms controlling their stability.
Figures
FIG. 1.
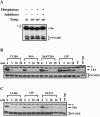
Abundance of Clb5 and Clb6 during the cell cycle. (A) Clb5 and Clb6 protein levels over time in a synchronous population of cells released from α-factor arrest. (B) Budding index and flow cytometric analysis of DNA content for the same time points. Times indicated are minutes after release from α-factor.
FIG. 2.
N-terminal deletions hyperstabilize Clb6. (A) ClustalW alignment of Clb5 and Clb6. Black boxes indicate identities. Gray boxes indicate putative phosphorylation sites (S/T-P) in Clb6. Clb5 D-box is shown. Arrows demarcate the start of Clb6 N-terminal deletions. (B) The relative stability of HA-tagged Clb6 N-terminal deletions. CLB6 transcription was induced from the GAL1 promoter by treating asynchronous cultures (in YEP-raffinose) with 0.1% galactose for 25 min. Cells were then harvested, and at t = 0 min, cells were resuspended in YEP-dextrose containing 1 mg/ml cycloheximide. Wild type (wt), peak levels of endogenous Clb6 (cells collected from synchronous population expressing CLB6-HA from its own promoter, 15 min after release from α-factor); no tag, cell lysates from cells expressing Clb6 without an HA epitope tag; PSTAIRE, loading control. The asterisk indicates an HA background band.
FIG. 3.
Clb6p hyperstabilized in _cdc4_-3 and _cdc34_-3 mutant cells at restrictive temperatures and in the presence of a proteosome inhibitor. (A and B) The relative stability of HA-tagged Clb6 in (A) wild-type (wt) cells versus _cdc4_-3 sic1Δ mutants at 33°C and (B) wild-type cells versus _cdc34_-3 sic1Δ mutants at 35°C. (C) The relative stability of HA-tagged Clb6 in wild-type cells versus sic1Δ cells. The promoter shutoff experiments whose results are shown in panels A and B were performed as described in the legend to Fig. 2 except that cultures were shifted to restrictive temperatures for 30 min and then treated with galactose at restrictive temperatures for 30 min as follows: wt/CLB6-HA cells, 0.2% galactose; _cdc4_-3 sic1Δ/CLB6-HA cells, 0.8% galactose; and _cdc34_-3 sic1Δ/CLB6 cells, 1% galactose. In panel C, cells were harvested from YEP-sucrose and grown in YEP-galactose for 30 min. Times indicated are minutes after resuspension in YEP-dextrose containing cycloheximide. wt, peak levels of endogenous Clb6. (D) Stability of HA-tagged Clb6 in cells treated or untreated with MG132. Mid-log-phase cultures in YEP-sucrose were treated with 50 μM MG132 in dimethyl sulfoxide (+) or dimethyl sulfoxide alone (−) and then grown at 30°C for 1 h. Galactose (0.5%) was then added to induce Clb6 expression from the GAL1 promoter for 30 min. Times indicated are minutes after resuspension in YEP-dextrose containing cycloheximide. (E) Slow-mobility forms of Clb6 appear in the presence of the proteasome inhibitor MG132 and the deubiquitinase inhibitor NEM. Cells expressing an HA-tagged Clb6 from the GAL1 promoter were untreated (−) or treated (+) with 100 μM MG132 in YEP-sucrose for 45 min. Clb6-HA expression was then induced by the addition of galactose for 45 min. Total protein was then isolated in the presence (+) or absence (−) of NEM. (D and E) Strains had the gene encoding the ABC transporter Pdr5 deleted to decrease the rate at which MG132 was pumped out of cells. PSTAIRE, loading control. The asterisk indicates an HA background band.
FIG. 4.
Clb6 is a phospho-protein and is hyperstabilized when putative phosphorylation sites are mutated to nonphosphorylatable residues. (A) Multiple species of Clb6 are observed in whole-cell lysates. Lysates were untreated (−) or treated (+) with lambda phosphatase (phosphatase) in the presence (+) or absence (−) of sodium fluoride and of sodium orthovanadate (inhibitors). The asterisk indicates slow-mobility bands. (B and C) The relative stability of wild-type Clb6 and Clb6 proteins bearing phosphorylation site mutations. The experiment was carried out as described in the legend to Fig. 2 except that cultures were treated with 0.05% galactose for 20 min. Clb6 (wt) and the S6A, S6AT39A, and Δ3P mutants are shown (B). Clb6 (wt) and the Δ3P and S147A mutants are shown (C). Times indicated are minutes after resuspending cells in YEP-dextrose containing cycloheximide. wt, peak levels of endogenous Clb6; no tag, cell lysates from cells expressing Clb6 without an HA epitope tag; PSTAIRE, loading control. The asterisk in panels B and C indicates an HA background band.
FIG. 5.
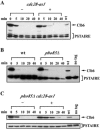
Both Cdc28 and Pho85 regulate the stability of Clb6. (A) Stability of Clb6-HA in cells expressing the analogue-sensitive allele cdc28-as1. Log-phase asynchronous cultures in YEP-sucrose were harvested, and Clb6-HA expression was induced in YEP-galactose medium for 45 min. Cultures were then either untreated (−) or treated (+) with 5 μM 1-NM-PP1 for 30 min. Times indicated are minutes after terminating expression by resuspending cells in YEP-dextrose containing cycloheximide. (B) Stability of Clb6-HA in cells with the PHO85 gene deleted. Cultures were treated as described in the legend to Fig. 2. (C) Stability of Clb6-HA in cdc28-as1 pho85Δ cells. cdc28-as1 pho85Δ cultures were either untreated (−) or treated (+) with 5 μM 1-NM-PP1 as described for panel A. wt, wild type; PSTAIRE, loading control.
FIG. 6.
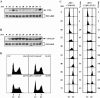
Clb6Δ3P expressed from the endogenous CLB6 promoter is hyperstable and suppresses the S-phase defect of clb5Δ cells. Wild-type Clb6 at its genomic locus was replaced by Clb6-HA or Clb6Δ3P-HA in clb5Δ cells. Clb6-HA (A) and Clb6Δ3P-HA (B) protein levels over time in a synchronous population of cells released from α-factor arrest. Times indicated are minutes after release from alpha factor. (C) Flow cytometric analysis of DNA content and budding index for the experiments whose results are shown in panels A and B. (D) Flow cytometric analysis of asynchronous, log-phase cultures. The percentage of cells in S phase is shown for each histogram. PSTAIRE, loading control.
FIG. 7.
Alignments of the N-terminal sequences of Clb6 for Saccharomyces species and the N-terminal sequences of A. gossypii. (A) The N-terminal amino acid sequence of Clb6 is shown for S. cerevisiae, S. bayanus, S. mikatae, and S. paradoxus. Sequences were downloaded from the Saccharomyces Genome Database (
http://db.yeastgenome.org/fungi/YGR109C.html
). Black boxes highlight putative S/TP phosphorylation sites. (B) The N-terminal amino acid sequence for AAR100C, the Clb5/Clb6 homolog from A. gossypii. The sequence was downloaded from the Ashbya Genome Database (
http://agd.unibas.ch/Ashbya\_gossypii/geneview?gene=AAR100C
). Black boxes highlight putative S/TP phosphorylation sites, and gray bars indicate D-box-like sequences.
Similar articles
- The anaphase-promoting complex works together with the SCF complex for proteolysis of the S-phase cyclin Clb6 during the transition from G1 to S phase.
Wu SY, Kuan VJ, Tzeng YW, Schuyler SC, Juang YL. Wu SY, et al. Fungal Genet Biol. 2016 Jun;91:6-19. doi: 10.1016/j.fgb.2016.03.004. Epub 2016 Mar 16. Fungal Genet Biol. 2016. PMID: 26994663 - S-phase cyclin-dependent kinases promote sister chromatid cohesion in budding yeast.
Hsu WS, Erickson SL, Tsai HJ, Andrews CA, Vas AC, Clarke DJ. Hsu WS, et al. Mol Cell Biol. 2011 Jun;31(12):2470-83. doi: 10.1128/MCB.05323-11. Epub 2011 Apr 25. Mol Cell Biol. 2011. PMID: 21518961 Free PMC article. - SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34, and cyclin/CDK activities.
Verma R, Feldman RM, Deshaies RJ. Verma R, et al. Mol Biol Cell. 1997 Aug;8(8):1427-37. doi: 10.1091/mbc.8.8.1427. Mol Biol Cell. 1997. PMID: 9285816 Free PMC article. - A process independent of the anaphase-promoting complex contributes to instability of the yeast S phase cyclin Clb5.
Sari F, Braus GH, Irniger S. Sari F, et al. J Biol Chem. 2007 Sep 7;282(36):26614-22. doi: 10.1074/jbc.M703744200. Epub 2007 Jul 9. J Biol Chem. 2007. PMID: 17620341 - Negative regulation of G1 and G2 by S-phase cyclins of Saccharomyces cerevisiae.
Basco RD, Segal MD, Reed SI. Basco RD, et al. Mol Cell Biol. 1995 Sep;15(9):5030-42. doi: 10.1128/MCB.15.9.5030. Mol Cell Biol. 1995. PMID: 7651421 Free PMC article.
Cited by
- A Mad2-Mediated Translational Regulatory Mechanism Promoting S-Phase Cyclin Synthesis Controls Origin Firing and Survival to Replication Stress.
Gay S, Piccini D, Bruhn C, Ricciardi S, Soffientini P, Carotenuto W, Biffo S, Foiani M. Gay S, et al. Mol Cell. 2018 May 17;70(4):628-638.e5. doi: 10.1016/j.molcel.2018.04.020. Mol Cell. 2018. PMID: 29775579 Free PMC article. - An overview of Cdk1-controlled targets and processes.
Enserink JM, Kolodner RD. Enserink JM, et al. Cell Div. 2010 May 13;5:11. doi: 10.1186/1747-1028-5-11. Cell Div. 2010. PMID: 20465793 Free PMC article. - Replication origins and timing of temporal replication in budding yeast: how to solve the conundrum?
Barberis M, Spiesser TW, Klipp E. Barberis M, et al. Curr Genomics. 2010 May;11(3):199-211. doi: 10.2174/138920210791110942. Curr Genomics. 2010. PMID: 21037857 Free PMC article. - Dual regulation by pairs of cyclin-dependent protein kinases and histone deacetylases controls G1 transcription in budding yeast.
Huang D, Kaluarachchi S, van Dyk D, Friesen H, Sopko R, Ye W, Bastajian N, Moffat J, Sassi H, Costanzo M, Andrews BJ. Huang D, et al. PLoS Biol. 2009 Sep;7(9):e1000188. doi: 10.1371/journal.pbio.1000188. Epub 2009 Sep 8. PLoS Biol. 2009. PMID: 19823668 Free PMC article. - The S-Phase Cyclin Clb5 Promotes rRNA Gene (rDNA) Stability by Maintaining Replication Initiation Efficiency in rDNA.
Goto M, Sasaki M, Kobayashi T. Goto M, et al. Mol Cell Biol. 2021 Apr 22;41(5):e00324-20. doi: 10.1128/MCB.00324-20. Print 2021 Apr 22. Mol Cell Biol. 2021. PMID: 33619126 Free PMC article.
References
- Bishop, A. C., J. A. Ubersax, D. T. Petsch, D. P. Matheos, N. S. Gray, J. Blethrow, E. Shimizu, J. Z. Tsien, P. G. Schultz, M. D. Rose, J. L. Wood, D. O. Morgan, and K. M. Shokat. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407:395-401. - PubMed
- Cliften, P., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton, J. Majors, R. Waterston, B. A. Cohen, and M. Johnston. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301:71-76. - PubMed
- Dietrich, F. S., S. Voegeli, S. Brachat, A. Lerch, K. Gates, S. Steiner, C. Mohr, R. Pohlmann, P. Luedi, S. Choi, R. A. Wing, A. Flavier, T. D. Gaffney, and P. Philippsen. 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304:304-307. - PubMed
- Donaldson, A. D., M. K. Raghuraman, K. L. Friedman, F. R. Cross, B. J. Brewer, and W. L. Fangman. 1998. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol. Cell 2:173-182. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials