PGC-1 coactivators: inducible regulators of energy metabolism in health and disease - PubMed (original) (raw)
Review
PGC-1 coactivators: inducible regulators of energy metabolism in health and disease
Brian N Finck et al. J Clin Invest. 2006 Mar.
Abstract
Members of the PPARgamma coactivator-1 (PGC-1) family of transcriptional coactivators serve as inducible coregulators of nuclear receptors in the control of cellular energy metabolic pathways. This Review focuses on the biologic and physiologic functions of the PGC-1 coactivators, with particular emphasis on striated muscle, liver, and other organ systems relevant to common diseases such as diabetes and heart failure.
Figures
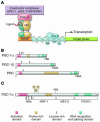
Figure 1
The PGC-1 coactivator family: inducible boosters of gene transcription. (A) The schematic uses generic NRs as an example of how inducible PGC-1 coactivators dock to transcription factor targets and recruit protein complexes that activate transcription via either enzymatic modification of chromatin, such as histone acetylation (e.g., by steroid receptor coactivator-1 [SRC-1] or p300), or direct interaction with the transcription initiation machinery (e.g., the thyroid hormone receptor–associated protein/vitamin D receptor–interacting protein [TRAP/DRIP] coactivator complex). The NR binds cognate NR response elements (NRREs) within the promoter region of the target gene. Specific histone modifications, including acetylation (Ac) and methylation (Me), are shown, as is the RNA polymerase II (Pol II) complex. (B) The schematic depicts the relative length and shared domains of the 3 members of the PGC-1 coactivator family. The nature of the domains is indicated in the key. (C) A schematic of the PGC-1α molecule is shown to denote several key functional domains involved in the interaction with specific target transcription factors including NRs, nuclear respiratory factor-1 (NRF-1), MEF-2, and FOXO1. MAPK phosphorylation (P) sites are also shown.
Figure 2
The PGC-1 gene regulatory cascade. The schematic indicates the upstream signaling events and downstream gene regulatory actions of the inducible PGC-1 coactivators, using PGC-1α as the representative factor. The interaction of PGC-1α with its cognate transcription factor targets is shown linked to specific organ systems. For example, PGC-1α coactivates members of the PPAR nuclear receptor transcription factor family, to activate the expression of genes involved in mitochondrial fatty acid oxidation. The signaling pathways shown at the top of each organ system transduce extracellular physiologic and nutritional stimuli to the expression and/or activity of PGC-1α. LXR, liver X receptor; TAG, triacylglycerol; RXR, retinoid X receptor; mtDNA, mitochondrial DNA; OXPHOS, oxidative phosphorylation.
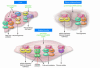
Figure 3
Potential contributions of organ-specific dysregulation of PGC-1α to the development of insulin resistance and type 2 diabetes. (A) PGC-1α expression and activity have been shown to be increased in the liver and pancreatic β cell in several animal models of diabetes mellitus. Conversely, gene expression profiling indicates that PGC-1α expression is diminished in skeletal muscle of type 1 and 2 diabetic humans along with reduced expression of genes involved in oxidative phosphorylation (OXPHOS). This tissue-specific pattern of dysregulated PGC-1α activity is predicted to potentially contribute to systemic insulin resistance, glucose intolerance, and insulin deficiency. (B) The generalized PGC-1α–deficient mouse is relatively protected against diet-induced insulin resistance and glucose intolerance despite impairments in skeletal muscle OXPHOS capacity. Improved insulin sensitivity may stem from diminished hepatic glucose production, a principal constituent of whole-body glucose homeostasis. However, the relative contribution of individual organ systems to the systemic insulin-sensitive phenotype requires further investigation.
Similar articles
- Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism.
Handschin C, Spiegelman BM. Handschin C, et al. Endocr Rev. 2006 Dec;27(7):728-35. doi: 10.1210/er.2006-0037. Epub 2006 Oct 3. Endocr Rev. 2006. PMID: 17018837 Review. - Intercellular: local and systemic actions of skeletal muscle PGC-1s.
Correia JC, Ferreira DM, Ruas JL. Correia JC, et al. Trends Endocrinol Metab. 2015 Jun;26(6):305-14. doi: 10.1016/j.tem.2015.03.010. Epub 2015 Apr 28. Trends Endocrinol Metab. 2015. PMID: 25934582 Review. - New insights into PGC-1 coactivators: redefining their role in the regulation of mitochondrial function and beyond.
Villena JA. Villena JA. FEBS J. 2015 Feb;282(4):647-72. doi: 10.1111/febs.13175. Epub 2015 Jan 12. FEBS J. 2015. PMID: 25495651 Review. - Transcriptional control of cardiac fuel metabolism and mitochondrial function.
Leone TC, Kelly DP. Leone TC, et al. Cold Spring Harb Symp Quant Biol. 2011;76:175-82. doi: 10.1101/sqb.2011.76.011965. Epub 2011 Nov 17. Cold Spring Harb Symp Quant Biol. 2011. PMID: 22096028 Free PMC article. Review. - PGC-1 coactivators in cardiac development and disease.
Rowe GC, Jiang A, Arany Z. Rowe GC, et al. Circ Res. 2010 Oct 1;107(7):825-38. doi: 10.1161/CIRCRESAHA.110.223818. Circ Res. 2010. PMID: 20884884 Free PMC article. Review.
Cited by
- Gene set of nuclear-encoded mitochondrial regulators is enriched for common inherited variation in obesity.
Knoll N, Jarick I, Volckmar AL, Klingenspor M, Illig T, Grallert H, Gieger C, Wichmann HE, Peters A, Hebebrand J, Scherag A, Hinney A. Knoll N, et al. PLoS One. 2013;8(2):e55884. doi: 10.1371/journal.pone.0055884. Epub 2013 Feb 8. PLoS One. 2013. PMID: 23409076 Free PMC article. - Regulation of the STARS signaling pathway in response to endurance and resistance exercise and training.
Lamon S, Wallace MA, Stefanetti RJ, Rahbek SK, Vendelbo MH, Russell AP, Vissing K. Lamon S, et al. Pflugers Arch. 2013 Sep;465(9):1317-25. doi: 10.1007/s00424-013-1265-5. Epub 2013 Mar 23. Pflugers Arch. 2013. PMID: 23525673 - Ventromedial hypothalamic melanocortin receptor activation: regulation of activity energy expenditure and skeletal muscle thermogenesis.
Gavini CK, Jones WC 2nd, Novak CM. Gavini CK, et al. J Physiol. 2016 Sep 15;594(18):5285-301. doi: 10.1113/JP272352. Epub 2016 May 29. J Physiol. 2016. PMID: 27126579 Free PMC article. - A highly integrated and complex PPARGC1A transcription factor binding network in HepG2 cells.
Charos AE, Reed BD, Raha D, Szekely AM, Weissman SM, Snyder M. Charos AE, et al. Genome Res. 2012 Sep;22(9):1668-79. doi: 10.1101/gr.127761.111. Genome Res. 2012. PMID: 22955979 Free PMC article. - Effects of two workload-matched high intensity interval training protocols on regulatory factors associated with mitochondrial biogenesis in the soleus muscle of diabetic rats.
Delfan M, Vahed A, Bishop DJ, Amadeh Juybari R, Laher I, Saeidi A, Granacher U, Zouhal H. Delfan M, et al. Front Physiol. 2022 Sep 23;13:927969. doi: 10.3389/fphys.2022.927969. eCollection 2022. Front Physiol. 2022. PMID: 36213227 Free PMC article.
References
- Puigserver P, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. - PubMed
- Lin J, Puigserver P, Donovan J, Tarr P, Spiegelman BM. Peroxisome proliferator-activated receptor γ coactivator 1β (PGC-1β), a novel PGC-1-related transcription coactivator associated with host cell factor. J. Biol. Chem. 2002;277:1645–1648. - PubMed
- Kressler D, Schreiber SN, Knutti D, Kralli A. The PGC-1-related protein PERC is a selective coactivator of estrogen receptor alpha. J. Biol. Chem. 2002;277:13918–13925. - PubMed
- Wu Z, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 HL57278/HL/NHLBI NIH HHS/United States
- R01 HL058493/HL/NHLBI NIH HHS/United States
- K01 DK062903/DK/NIDDK NIH HHS/United States
- P30 DK056341/DK/NIDDK NIH HHS/United States
- P30 DK56341/DK/NIDDK NIH HHS/United States
- R01 DK045416/DK/NIDDK NIH HHS/United States
- P30 DK056341-06/DK/NIDDK NIH HHS/United States
- R01 HL58493/HL/NHLBI NIH HHS/United States
- P50 HL077113/HL/NHLBI NIH HHS/United States
- P30 DK056341-05S2/DK/NIDDK NIH HHS/United States
- P30 DK52574/DK/NIDDK NIH HHS/United States
- P01 HL057278/HL/NHLBI NIH HHS/United States
- P30 DK052574/DK/NIDDK NIH HHS/United States
- R01 DK45416/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases