An improved map of conserved regulatory sites for Saccharomyces cerevisiae - PubMed (original) (raw)
An improved map of conserved regulatory sites for Saccharomyces cerevisiae
Kenzie D MacIsaac et al. BMC Bioinformatics. 2006.
Abstract
Background: The regulatory map of a genome consists of the binding sites for proteins that determine the transcription of nearby genes. An initial regulatory map for S. cerevisiae was recently published using six motif discovery programs to analyze genome-wide chromatin immunoprecipitation data for 203 transcription factors. The programs were used to identify sequence motifs that were likely to correspond to the DNA-binding specificity of the immunoprecipitated proteins. We report improved versions of two conservation-based motif discovery algorithms, PhyloCon and Converge. Using these programs, we create a refined regulatory map for S. cerevisiae by reanalyzing the same chromatin immunoprecipitation data.
Results: Applying the same conservative criteria that were applied in the original study, we find that PhyloCon and Converge each separately discover more known specificities than the combination of all six programs in the previous study. Combining the results of PhyloCon and Converge, we discover significant sequence motifs for 36 transcription factors that were previously missed. The new set of motifs identifies 636 more regulatory interactions than the previous one. The new network contains 28% more regulatory interactions among transcription factors, evidence of greater cross-talk between regulators.
Conclusion: Combining two complementary computational strategies for conservation-based motif discovery improves the ability to identify the specificity of transcriptional regulators from genome-wide chromatin immunoprecipitation data. The increased sensitivity of these methods significantly expands the map of yeast regulatory sites without the need to alter any of the thresholds for statistical significance. The new map of regulatory sites reveals a more elaborate and complex view of the yeast genetic regulatory network than was observed previously.
Figures
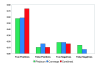
Figure 1
Performance of PhyloCon, Converge, and the combined motif set on data for factors of known specificity. PhyloCon and Converge both recover more true positives than the suite of 6 programs employed in Harbison et al. Combining the results of PhyloCon and Converge significantly increases the number of true positives recovered, and eliminates false negatives, without a large adverse effect on the false positive rate. For definitions of the scoring criteria, see the Methods section.
Figure 2
Selected Factor Specificities in the New Yeast Regulatory Map.
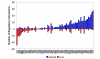
Figure 3
Changes in the number of putative regulatory interactions for factors common to the old and new regulatory codes. For each modified motif, the number of regulatory interactions added and lost relative to the previously reported map is shown. Our analysis produced modified factor binding specificities for 85 factors, resulting in a net gain of 398 putatively regulated genes.
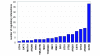
Figure 4
Regulatory interactions added through the addition of new factor specificity estimates. A total of 200 genes were identified as being putatively regulated by factors with newly reported motifs.
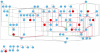
Figure 5
Yeast transcriptional regulatory network. Nodes correspond to transcription factors and an edge from one factor to another indicates that the first factor regulates the second. Red nodes correspond to factors without a previously reported specificity. Edges are colored red for interactions unique to the new map, grey for interactions common to the old and new maps, and green for interactions unique to the old map. There are 39 new interactions gained and 6 interactions lost relative to the previous map.
Similar articles
- Phylogeny based discovery of regulatory elements.
Gertz J, Fay JC, Cohen BA. Gertz J, et al. BMC Bioinformatics. 2006 May 22;7:266. doi: 10.1186/1471-2105-7-266. BMC Bioinformatics. 2006. PMID: 16716228 Free PMC article. - Discriminative discovery of transcription factor binding sites from location data.
Kawada Y, Sakakibara Y. Kawada Y, et al. Proc IEEE Comput Syst Bioinform Conf. 2005:86-9. doi: 10.1109/csb.2005.30. Proc IEEE Comput Syst Bioinform Conf. 2005. PMID: 16447966 - Identifying the conserved network of cis-regulatory sites of a eukaryotic genome.
Wang T, Stormo GD. Wang T, et al. Proc Natl Acad Sci U S A. 2005 Nov 29;102(48):17400-5. doi: 10.1073/pnas.0505147102. Epub 2005 Nov 21. Proc Natl Acad Sci U S A. 2005. PMID: 16301543 Free PMC article. - Finding regulatory elements and regulatory motifs: a general probabilistic framework.
van Nimwegen E. van Nimwegen E. BMC Bioinformatics. 2007 Sep 27;8 Suppl 6(Suppl 6):S4. doi: 10.1186/1471-2105-8-S6-S4. BMC Bioinformatics. 2007. PMID: 17903285 Free PMC article. Review.
Cited by
- Assigning roles to DNA regulatory motifs using comparative genomics.
Buske FA, Bodén M, Bauer DC, Bailey TL. Buske FA, et al. Bioinformatics. 2010 Apr 1;26(7):860-6. doi: 10.1093/bioinformatics/btq049. Epub 2010 Feb 10. Bioinformatics. 2010. PMID: 20147307 Free PMC article. - High-resolution transcription atlas of the mitotic cell cycle in budding yeast.
Granovskaia MV, Jensen LJ, Ritchie ME, Toedling J, Ning Y, Bork P, Huber W, Steinmetz LM. Granovskaia MV, et al. Genome Biol. 2010;11(3):R24. doi: 10.1186/gb-2010-11-3-r24. Epub 2010 Mar 1. Genome Biol. 2010. PMID: 20193063 Free PMC article. - H1.0 C Terminal Domain Is Integral for Altering Transcription Factor Binding within Nucleosomes.
Burge NL, Thuma JL, Hong ZZ, Jamison KB, Ottesen JJ, Poirier MG. Burge NL, et al. Biochemistry. 2022 Apr 19;61(8):625-638. doi: 10.1021/acs.biochem.2c00001. Epub 2022 Apr 4. Biochemistry. 2022. PMID: 35377618 Free PMC article. - YPA: an integrated repository of promoter features in Saccharomyces cerevisiae.
Chang DT, Huang CY, Wu CY, Wu WS. Chang DT, et al. Nucleic Acids Res. 2011 Jan;39(Database issue):D647-52. doi: 10.1093/nar/gkq1086. Epub 2010 Nov 2. Nucleic Acids Res. 2011. PMID: 21045055 Free PMC article. - Is transcription factor binding site turnover a sufficient explanation for cis-regulatory sequence divergence?
Venkataram S, Fay JC. Venkataram S, et al. Genome Biol Evol. 2010;2:851-8. doi: 10.1093/gbe/evq066. Epub 2010 Nov 10. Genome Biol Evol. 2010. PMID: 21068212 Free PMC article.
References
- Long F, Liu H, Hahn C, Sumazin P, Zhang MQ, Zilberstein A. Genome-wide prediction and analysis of function-specific transcription factor binding sites. In Silico Biol. 2004;4:395–410. - PubMed
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. - DOI - PMC - PubMed
- Moses AM, Chiang DY, Eisen MB. Phylogenetic motif detection by expectation-maximization on evolutionary mixtures. Pac Symp Biocomput. 2004:324–335. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 1R01 HG002668-01/HG/NHGRI NIH HHS/United States
- 2T32HG00045/HG/NHGRI NIH HHS/United States
- T32 HG000045/HG/NHGRI NIH HHS/United States
- R01 HG000249/HG/NHGRI NIH HHS/United States
- HG00249/HG/NHGRI NIH HHS/United States
- R01 HG002668/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases