Genome-wide analysis of re-replication reveals inhibitory controls that target multiple stages of replication initiation - PubMed (original) (raw)
Genome-wide analysis of re-replication reveals inhibitory controls that target multiple stages of replication initiation
Robyn E Tanny et al. Mol Biol Cell. 2006 May.
Abstract
DNA replication must be tightly controlled during each cell cycle to prevent unscheduled replication and ensure proper genome maintenance. The currently known controls that prevent re-replication act redundantly to inhibit pre-replicative complex (pre-RC) assembly outside of the G1-phase of the cell cycle. The yeast Saccharomyces cerevisiae has been a useful model organism to study how eukaryotic cells prevent replication origins from reinitiating during a single cell cycle. Using a re-replication-sensitive strain and DNA microarrays, we map sites across the S. cerevisiae genome that are re-replicated as well as sites of pre-RC formation during re-replication. Only a fraction of the genome is re-replicated by a subset of origins, some of which are capable of multiple reinitiation events. Translocation experiments demonstrate that origin-proximal sequences are sufficient to predispose an origin to re-replication. Origins that reinitiate are largely limited to those that can recruit Mcm2-7 under re-replicating conditions; however, the formation of a pre-RC is not sufficient for reinitiation. Our findings allow us to categorize origins with respect to their propensity to reinitiate and demonstrate that pre-RC formation is not the only target for the mechanisms that prevent genomic re-replication.
Figures
Figure 1.
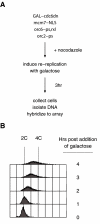
Multiple pre-RC mutations result in induced re-replication. (A) An outline of the re-replication experiment. Re-replication-sensitive cells were grown to an OD600 of 0.4 in YPD and then arrested in nocodazole. After the cells were arrested, galactose was added to induce expression of Cdc6ΔN. After 3 h, cells were collected for further experiments. (B) FACS analysis of the re-replication-sensitive strain SB1507 (See Supplementary Table 2) several hours after induction of re-replication.
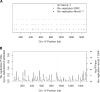
Figure 2.
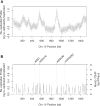
Analysis of genome-wide re-replication. (A) Re-replication is detected by copy number analysis using DNA microarrays. DNA from re-replicating cells and from G1-arrested cells was differentially labeled and cohybridized to a high-density DNA microarray. The log ratio re-replicated/unreplicated for each spot was plotted as a function of its position along the chromosome. Chromosome IV (see Supplementary Figure 2 for other chromosomes) is shown here as an example. (B) Sites of re-replication initiation are associated with G1 Mcm2-7 binding sites. A smoothing algorithm and a significance cutoff was applied to the re-replication data (see Materials and Methods) and plotted here for Chromosome IV (gray histogram). G1 Mcm2-7 binding sites (black histogram), determined by genome-wide location analysis, are superimposed on top of the re-replication data. Key sites discussed throughout the text, ARS1, ARS418, ARS428, and iYDR309C, are marked with a gray dashed line.
Figure 3.
An origin sequence directs reinitiation (A) The re-replication profile surrounding iYDR309C, a segment that does not re-replicate, in the absence of an ectopic origin is depicted as both the gray histogram and the black dashed line. (B) ARS418, an origin that is associated with a peak on the re-replication profile, directs re-replication at an ectopic locus. The 600-base pair intergenic region containing ARS418 was moved to iYDR309C. Re-replication was induced and the resulting DNA was hybridized to a low-density DNA microarray (gray histogram, Chromosome IV, 800-1400 kb). Superimposed on top is the re-replication profile from the strain without the ectopic ARS418 (black dashed line). (C) An origin with a mutant ACS is not capable of directing re-replication. The essential ACS of ARS418 was mutated and integrated into the re-replicating strain. Re-replication was induced and the resulting DNA hybridized on to a low-density array (gray histogram, Chromosome IV, 800-1400 kb). The re-replication profile of the strain without the ectopic mutant ARS418 is superimposed on top (black dashed line).
Figure 4.
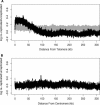
Subtelomeric regions have a high probability of re-replicating. (A) There is a positive correlation between the proximity of a sequence to the telomere and its probability of re-replicating. The relative enrichment for each spot on the microarray was plotted as a function of its distance to the closest telomere for both the re-replicating strain (black) and wild-type strain (gray) 3 h after addition of galactose. (B) There is no correlation between re-replication and proximity to centromeres. The relative enrichment for each spot on the microarray was plotted as a function of its distance to the centromere for both the re-replicating strain (black) and wild-type strain (gray) 3 h after addition of galactose.
Figure 5.
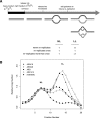
Origins are capable of reinitiating multiple times. (A) Diagram of density transfer experiment. A cartoon depicts what products will look like during the experiment with “heavy” DNA strands shown in black and “light” DNA strands shown in gray. The table briefly describes possible results. (B) ARS428 and ARS418 reinitiate multiple times. DNA from cells that underwent the density transfer protocol described in A was fractionated by CsCl gradient. The resulting fractions were probed for three different classes of DNA sequences as determined by DNA microarray analysis: two origins of reinitiation (ARS418, ▴; ARS428, □), two origins that are re-replicated, but are not sites of reinitiation (ARS1, ▪; ARS1413, ○) and an intergenic sequence that does not re-replicate (iYDR309C, •). The data were normalized by setting the peak of the HL density to a copy number of one.
Figure 6.
Recruitment of Mcm2-7 is not sufficient for reinitiation. (A) Mcm2-7 binds only a fraction of possible origins during re-replication. Genome-wide location analysis of Mcm2-7 and ORC was performed 45 min after induction of re-replication. The binding sites of Mcm2-7 during re-replication (•) were compared with binding sites of ORC during re-replication (dark gray circles) and binding sites of Mcm2-7 during G1 (light gray circles). Plotted are only the points on the array that satisfied the significance cutoff (see Materials and Methods) for each of the data sets. (B) Mcm2-7 binds to origins that do not reinitiate. The binding sites of Mcm2-7 (black histogram) are overlaid on top of the re-replication profile for Chromosome IV (gray histogram). Each peak of re-replication is associated with an Mcm2-7 binding site, but the converse is not true.
Similar articles
- Genome-wide mapping of DNA synthesis in Saccharomyces cerevisiae reveals that mechanisms preventing reinitiation of DNA replication are not redundant.
Green BM, Morreale RJ, Ozaydin B, Derisi JL, Li JJ. Green BM, et al. Mol Biol Cell. 2006 May;17(5):2401-14. doi: 10.1091/mbc.e05-11-1043. Epub 2006 Feb 15. Mol Biol Cell. 2006. PMID: 16481397 Free PMC article. - Nucleosome occupancy as a novel chromatin parameter for replication origin functions.
Rodriguez J, Lee L, Lynch B, Tsukiyama T. Rodriguez J, et al. Genome Res. 2017 Feb;27(2):269-277. doi: 10.1101/gr.209940.116. Epub 2016 Nov 28. Genome Res. 2017. PMID: 27895110 Free PMC article. - Regulatory mechanisms that prevent re-initiation of DNA replication can be locally modulated at origins by nearby sequence elements.
Richardson CD, Li JJ. Richardson CD, et al. PLoS Genet. 2014 Jun 19;10(6):e1004358. doi: 10.1371/journal.pgen.1004358. eCollection 2014 Jun. PLoS Genet. 2014. PMID: 24945837 Free PMC article. - Surveying genome replication.
Kearsey S. Kearsey S. Genome Biol. 2002;3(6):REVIEWS1016. doi: 10.1186/gb-2002-3-6-reviews1016. Epub 2002 May 24. Genome Biol. 2002. PMID: 12093380 Free PMC article. Review. - Cell cycle control and initiation of DNA replication in Saccharomyces cerevisiae.
Küntzel H, Schulz A, Ehbrecht IM. Küntzel H, et al. Biol Chem. 1996 Jul-Aug;377(7-8):481-7. Biol Chem. 1996. PMID: 8922282 Review.
Cited by
- RAD51 restricts DNA over-replication from re-activated origins.
Muñoz S, Blanco-Romero E, González-Acosta D, Rodriguez-Acebes S, Megías D, Lopes M, Méndez J. Muñoz S, et al. EMBO J. 2024 Mar;43(6):1043-1064. doi: 10.1038/s44318-024-00038-z. Epub 2024 Feb 15. EMBO J. 2024. PMID: 38360996 Free PMC article. - Unscheduled DNA replication in G1 causes genome instability and damage signatures indicative of replication collisions.
Reusswig KU, Bittmann J, Peritore M, Courtes M, Pardo B, Wierer M, Mann M, Pfander B. Reusswig KU, et al. Nat Commun. 2022 Nov 18;13(1):7014. doi: 10.1038/s41467-022-34379-2. Nat Commun. 2022. PMID: 36400763 Free PMC article. - In silico analysis of DNA re-replication across a complete genome reveals cell-to-cell heterogeneity and genome plasticity.
Rapsomaniki MA, Maxouri S, Nathanailidou P, Garrastacho MR, Giakoumakis NN, Taraviras S, Lygeros J, Lygerou Z. Rapsomaniki MA, et al. NAR Genom Bioinform. 2021 Jan 28;3(1):lqaa112. doi: 10.1093/nargab/lqaa112. eCollection 2021 Mar. NAR Genom Bioinform. 2021. PMID: 33554116 Free PMC article. - DNA copy-number measurement of genome replication dynamics by high-throughput sequencing: the sort-seq, sync-seq and MFA-seq family.
Batrakou DG, Müller CA, Wilson RHC, Nieduszynski CA. Batrakou DG, et al. Nat Protoc. 2020 Mar;15(3):1255-1284. doi: 10.1038/s41596-019-0287-7. Epub 2020 Feb 12. Nat Protoc. 2020. PMID: 32051615 - Bayesian inference of origin firing time distributions, origin interference and licencing probabilities from Next Generation Sequencing data.
Bazarova A, Nieduszynski CA, Akerman I, Burroughs NJ. Bazarova A, et al. Nucleic Acids Res. 2019 Mar 18;47(5):2229-2243. doi: 10.1093/nar/gkz094. Nucleic Acids Res. 2019. PMID: 30859196 Free PMC article.
References
- Aparicio, O. M., Weinstein, D. M., and Bell, S. P. (1997). Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91, 59-69. - PubMed
- Bell, S. P., and Dutta, A. (2002). DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333-374. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous