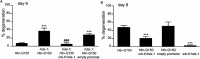Differential contributions of Caenorhabditis elegans histone deacetylases to huntingtin polyglutamine toxicity - PubMed (original) (raw)
Comparative Study
Differential contributions of Caenorhabditis elegans histone deacetylases to huntingtin polyglutamine toxicity
Emily A Bates et al. J Neurosci. 2006.
Abstract
Expansion of a polyglutamine tract in the huntingtin protein causes neuronal degeneration and death in Huntington's disease patients, but the molecular mechanisms underlying polyglutamine-mediated cell death remain unclear. Previous studies suggest that expanded polyglutamine tracts alter transcription by sequestering glutamine rich transcriptional regulatory proteins, thereby perturbing their function. We tested this hypothesis in Caenorhabditis elegans neurons expressing a human huntingtin fragment with an expanded polyglutamine tract (Htn-Q150). Loss of function alleles and RNA interference (RNAi) were used to examine contributions of C. elegans cAMP response element-binding protein (CREB), CREB binding protein (CBP), and histone deacetylases (HDACs) to polyglutamine-induced neurodegeneration. Deletion of CREB (crh-1) or loss of one copy of CBP (cbp-1) enhanced polyglutamine toxicity in C. elegans neurons. Loss of function alleles and RNAi were then used to systematically reduce function of each C. elegans HDAC. Generally, knockdown of individual C. elegans HDACs enhanced Htn-Q150 toxicity, but knockdown of C. elegans hda-3 suppressed toxicity. Neuronal expression of hda-3 restored Htn-Q150 toxicity and suggested that C. elegans HDAC3 (HDA-3) acts within neurons to promote degeneration in response to Htn-Q150. Genetic epistasis experiments suggested that HDA-3 and CRH-1 (C. elegans CREB homolog) directly oppose each other in regulating transcription of genes involved in polyglutamine toxicity. hda-3 loss of function failed to suppress increased neurodegeneration in hda-1/+;Htn-Q150 animals, indicating that HDA-1 and HDA-3 have different targets with opposing effects on polyglutamine toxicity. Our results suggest that polyglutamine expansions perturb transcription of CREB/CBP targets and that specific targeting of HDACs will be useful in reducing associated neurodegeneration.
Figures
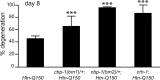
Figure 1.
cbp-1(ys2)/+, cbp-1(ys4)/+, and_crh-1(3315)_ enhance Htn-Q150-induced degeneration. Sibling controls for cbp-1(ys2)/+; Htn-Q150,cbp-1(ys4)/+;Htn-Q150, and crh-1(e3315); Htn-Q150 were combined, because degeneration levels were not significantly different. No degeneration was detected in_cbp-1(ys2)/+, cbp-1(ys4)/+, or_crh-1 animals that did not express Htn-Q150. Error bars represent SD. ***p < 0.005 versus column 1. At least three trials were conducted for each experiment to total _n_ > 150.
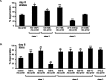
Figure 2.
A, hda-1(e1795)/+, hda-4, sir-2.1(ok434) enhance Htn-Q150 toxicity. sir-2.1_overexpression suppresses Htn-Q150 toxicity. sir-2.3 has no effect on Htn-Q150 toxicity. Controls were combined, because they are not significantly different. No degeneration was detected in hda-1(e1795)/+,hda-4, sir-2.1(ok434), or sir-2.3 animals without expression of Htn-Q150. At least three trials were conducted for each experiment to total n > 150. B, RNAi of class I HDACs, hda-1 and hda-2, enhances Htn-Q150 toxicity. RNAi of class I HDAC, hda-3, suppresses Htn-Q150 toxicity. RNAi of class II HDACs enhanced degeneration, as did RNAi of_sir-2.2, a class III HDAC. Controls were combined, because they are not significantly different. At least three trials were conducted for each experiment to total n > 150. Error bars represent SD. *p < 0.05, **p < 0.005, ***p < 0.0005 in pairwise comparison with column 1 using a two-tailed t test.
Figure 3.
A, Expression of HDA-1 in ASH neurons restores Htn-Q150 toxicity to normal levels. Injection of equal concentrations of an empty_srb-6_ promoter plasmid and the transgenic marker fail to rescue the hda-1(e1795) enhancement of polyQ toxicity (29 ± 2%).B, In Htn-Q150 animals without the_srb-6::hda-1_ transgene, 47 ± 4% of the ASH neurons degenerate by day 8. Degeneration is decreased to 20 ± 4% by increased expression of HDA-1 (50 ng/μl) in Htn-Q150 ASH neurons. Expression of HDA-1 does not cause detectible degeneration of ASH neurons that do not express Htn-Q150 (2 ± 2%). No significant change in neurodegeneration is detectable in animals that carry an empty srb-6 promoter and the transgenic marker_myo-2_::GFP (50 ± 10%). ***p < 0.0005 compared with corresponding _Htn-Q150_ animals in column 1;###_p_ < 0.0005 when comparing_hda-1;Htn-Q150; srb-65::hda-1_ with_hda-1;Htn-Q150_ in column 2 (to assess rescue) in**_A_**. Empty promoter transgenes did not significantly change polyglutamine toxicity. At least three trials were conducted for each experiment to total _n_ > 150. Error bars represent SD.
Figure 4.
HDA-3 specifically rescues hda-3(tm1374) suppression of Htn-Q150 in ASH neurons. Degeneration is reduced from 27 ± 2% in_rtIs11_Htn-Q150 animals to 6 ± 6% in hda-3(tm1374); rtIs11(Htn-Q150) animals at day 7. Degeneration ranges from 14 to 63%, depending on the transgenic line (average, 45 ± 22%) in hda-3(tm1374); rtIs11(Htn-Q150); srb-6:hda-3 transgenic animals. As a control, a plasmid carrying the empty srb-6 promoter was injected along with transgenic markers. The empty promoter has no significant effect on polyQ toxicity (10 ± 3%). Overexpression of hda-3 in ASH using the_srb-6_ promoter increases degeneration from 24 ± 11 to 45 ± 13% (p = 0.03). ***p < 0.0005 compared with column 1 for each graph; ###_p_ < 0.0005 when comparing _hda-3;Htn-Q150; srb-6::hda3_ with_hda-3;Htn-Q150_. At least three trials were conducted for each experiment to total _n_ > 150. Error bars represent SD.
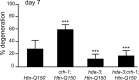
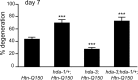
Figure 5.
HDA-3 modulates Htn-Q150 toxicity via transcriptional regulation. Loss of_hda-3_ function suppresses the deleterious effects of loss of_crh-1_ function. ASH neuron degeneration is increased in_crh-1;rtIs11[Htn-Q150]_ animals and decreased in_hda-3(tm1374);rtIs11[Htn-Q150]_ animals compared with_rtIs11[Htn-Q150]. ASH degeneration is suppressed in_hda-3(tm1374);crh-1; rtIs11[Htn-Q150] animals and is not significantly different from degeneration levels in_hda-3(tm1374);rtIs11[Htn-Q150]_ animals at day 7. Error bars represent SD. ***p < 0.0005 compared with column 1 of each graph. At least three trials were conducted for each determination to total_n_ > 150.
Figure 6.
Normal hda-1 levels are critical for hda-3_modulation of polyglutamine toxicity. Loss of one copy of hda-1_eliminates the salubrious effects of hda-3 loss of function. ASH neuron degeneration is increased in hda-1(e1795)/+; rtIs14[Htn-Q150] animals and decreased in_hda-3(tm1374);rtIs14[Htn-Q150i] animals compared with_rtIs14[Htn-Q150] animals. Degeneration levels are increased in_hda-3(tm1374);hda-1(e1795)/+; rtIs14[Htn-Q150]_ and are not significantly different from degeneration levels in hda-1(e1795)/+; rtIs14[Htn-Q150] animals. Error bars represent SD. ***p< 0.0005 compared with column 1. At least three trials were conducted for each determination to total _n_ > 150.
Similar articles
- Polyglutamine-mediated dysfunction and apoptotic death of a Caenorhabditis elegans sensory neuron.
Faber PW, Alter JR, MacDonald ME, Hart AC. Faber PW, et al. Proc Natl Acad Sci U S A. 1999 Jan 5;96(1):179-84. doi: 10.1073/pnas.96.1.179. Proc Natl Acad Sci U S A. 1999. PMID: 9874792 Free PMC article. - Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila.
Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL, Kazantsev A, Schmidt E, Zhu YZ, Greenwald M, Kurokawa R, Housman DE, Jackson GR, Marsh JL, Thompson LM. Steffan JS, et al. Nature. 2001 Oct 18;413(6857):739-43. doi: 10.1038/35099568. Nature. 2001. PMID: 11607033 - Depletion of CBP is directly linked with cellular toxicity caused by mutant huntingtin.
Jiang H, Poirier MA, Liang Y, Pei Z, Weiskittel CE, Smith WW, DeFranco DB, Ross CA. Jiang H, et al. Neurobiol Dis. 2006 Sep;23(3):543-51. doi: 10.1016/j.nbd.2006.04.011. Neurobiol Dis. 2006. PMID: 16766198 - Nucleocytoplasmic trafficking and transcription effects of huntingtin in Huntington's disease.
Truant R, Atwal RS, Burtnik A. Truant R, et al. Prog Neurobiol. 2007 Nov;83(4):211-27. doi: 10.1016/j.pneurobio.2006.11.004. Epub 2007 Jan 22. Prog Neurobiol. 2007. PMID: 17240517 Review. - Genetic and pharmacological suppression of polyglutamine-dependent neuronal dysfunction in Caenorhabditis elegans.
Parker JA, Holbert S, Lambert E, Abderrahmane S, Néri C. Parker JA, et al. J Mol Neurosci. 2004;23(1-2):61-8. doi: 10.1385/JMN:23:1-2:061. J Mol Neurosci. 2004. PMID: 15126693 Review.
Cited by
- Huntington's disease: the coming of age.
Pandey M, Rajamma U. Pandey M, et al. J Genet. 2018 Jul;97(3):649-664. J Genet. 2018. PMID: 30027901 Review. - Inhibition of specific HDACs and sirtuins suppresses pathogenesis in a Drosophila model of Huntington's disease.
Pallos J, Bodai L, Lukacsovich T, Purcell JM, Steffan JS, Thompson LM, Marsh JL. Pallos J, et al. Hum Mol Genet. 2008 Dec 1;17(23):3767-75. doi: 10.1093/hmg/ddn273. Epub 2008 Sep 1. Hum Mol Genet. 2008. PMID: 18762557 Free PMC article. - Acetylation targets mutant huntingtin to autophagosomes for degradation.
Jeong H, Then F, Melia TJ Jr, Mazzulli JR, Cui L, Savas JN, Voisine C, Paganetti P, Tanese N, Hart AC, Yamamoto A, Krainc D. Jeong H, et al. Cell. 2009 Apr 3;137(1):60-72. doi: 10.1016/j.cell.2009.03.018. Cell. 2009. PMID: 19345187 Free PMC article. - Axon regeneration pathways identified by systematic genetic screening in C. elegans.
Chen L, Wang Z, Ghosh-Roy A, Hubert T, Yan D, O'Rourke S, Bowerman B, Wu Z, Jin Y, Chisholm AD. Chen L, et al. Neuron. 2011 Sep 22;71(6):1043-57. doi: 10.1016/j.neuron.2011.07.009. Epub 2011 Sep 21. Neuron. 2011. PMID: 21943602 Free PMC article. - Disassociation of histone deacetylase-3 from normal huntingtin underlies mutant huntingtin neurotoxicity.
Bardai FH, Verma P, Smith C, Rawat V, Wang L, D'Mello SR. Bardai FH, et al. J Neurosci. 2013 Jul 17;33(29):11833-8. doi: 10.1523/JNEUROSCI.5831-12.2013. J Neurosci. 2013. PMID: 23864673 Free PMC article.
References
- Ajamian F, Salminen A, Reeben M (2004). Selective regulation of class I and class II histone deacetylases expression by inhibitors of histone deacetylases in cultured mouse neural cells. Neurosci Lett 365:64–68. - PubMed
- Baran R, Aronoff R, Garriga G (1999). The C. elegans homeodomain gene unc-42 regulates chemosensory and glutamate receptor expression. Development 126:2241–2251. - PubMed
- Boutell JM, Thomas P, Neal JW, Weston VJ, Duce J, Harper PS, Jones AL (1999). Aberrant interactions of transcriptional repressor proteins with the Huntington's disease gene product, huntingtin. Hum Mol Genet 8:1647–1655. - PubMed
- Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD (1995). The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev 9:2888–2902. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous