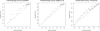A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase - PubMed (original) (raw)
A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase
Paul Scheet et al. Am J Hum Genet. 2006 Apr.
Abstract
We present a statistical model for patterns of genetic variation in samples of unrelated individuals from natural populations. This model is based on the idea that, over short regions, haplotypes in a population tend to cluster into groups of similar haplotypes. To capture the fact that, because of recombination, this clustering tends to be local in nature, our model allows cluster memberships to change continuously along the chromosome according to a hidden Markov model. This approach is flexible, allowing for both "block-like" patterns of linkage disequilibrium (LD) and gradual decline in LD with distance. The resulting model is also fast and, as a result, is practicable for large data sets (e.g., thousands of individuals typed at hundreds of thousands of markers). We illustrate the utility of the model by applying it to dense single-nucleotide-polymorphism genotype data for the tasks of imputing missing genotypes and estimating haplotypic phase. For imputing missing genotypes, methods based on this model are as accurate or more accurate than existing methods. For haplotype estimation, the point estimates are slightly less accurate than those from the best existing methods (e.g., for unrelated Centre d'Etude du Polymorphisme Humain individuals from the HapMap project, switch error was 0.055 for our method vs. 0.051 for PHASE) but require a small fraction of the computational cost. In addition, we demonstrate that the model accurately reflects uncertainty in its estimates, in that probabilities computed using the model are approximately well calibrated. The methods described in this article are implemented in a software package, fastPHASE, which is available from the Stephens Lab Web site.
Figures
Figure 1
Illustration of how our model allows cluster membership to change continuously along a chromosome. Each column represents a SNP, with the two alleles indicated by open and crossed squares. Successive pairs of rows represent the estimated pair of haplotypes for successive individuals. Colors represent estimated cluster membership of each allele, which changes as one moves along each haplotype. Locally, each cluster can be thought of as representing a (common) combination of alleles at tightly linked SNPs, and the figure illustrates how each haplotype is modeled as a mosaic of these common combinations. The figure was produced by fitting our model to the HapMap data from 60 unrelated CEPH individuals (see the “Results” section) and then taking a single sample of cluster memberships and haplotypes from their conditional distribution, given the genotype data and parameter estimates (appendix B). For brevity, haplotypes from only 10 individuals are shown.
Figure 2
Calibration of our model for predicting uncertainty in inferred genotypes and haplotypes. Points (triangles) represent probabilities obtained by averaging over the 20 runs of the EM algorithm, as described in the text.
Similar articles
- Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation.
Stephens M, Scheet P. Stephens M, et al. Am J Hum Genet. 2005 Mar;76(3):449-62. doi: 10.1086/428594. Epub 2005 Jan 31. Am J Hum Genet. 2005. PMID: 15700229 Free PMC article. - Inference of haplotypic phase and missing genotypes in polyploid organisms and variable copy number genomic regions.
Su SY, White J, Balding DJ, Coin LJ. Su SY, et al. BMC Bioinformatics. 2008 Dec 1;9:513. doi: 10.1186/1471-2105-9-513. BMC Bioinformatics. 2008. PMID: 19046436 Free PMC article. - A comparison of phasing algorithms for trios and unrelated individuals.
Marchini J, Cutler D, Patterson N, Stephens M, Eskin E, Halperin E, Lin S, Qin ZS, Munro HM, Abecasis GR, Donnelly P; International HapMap Consortium. Marchini J, et al. Am J Hum Genet. 2006 Mar;78(3):437-50. doi: 10.1086/500808. Epub 2006 Jan 26. Am J Hum Genet. 2006. PMID: 16465620 Free PMC article. - Inferring coancestry in population samples in the presence of linkage disequilibrium.
Brown MD, Glazner CG, Zheng C, Thompson EA. Brown MD, et al. Genetics. 2012 Apr;190(4):1447-60. doi: 10.1534/genetics.111.137570. Epub 2012 Jan 31. Genetics. 2012. PMID: 22298700 Free PMC article. - Linkage disequilibrium-based quality control for large-scale genetic studies.
Scheet P, Stephens M. Scheet P, et al. PLoS Genet. 2008 Aug 1;4(8):e1000147. doi: 10.1371/journal.pgen.1000147. PLoS Genet. 2008. PMID: 18670630 Free PMC article.
Cited by
- Assessment of the genomic variation in a cattle population by re-sequencing of key animals at low to medium coverage.
Jansen S, Aigner B, Pausch H, Wysocki M, Eck S, Benet-Pagès A, Graf E, Wieland T, Strom TM, Meitinger T, Fries R. Jansen S, et al. BMC Genomics. 2013 Jul 4;14:446. doi: 10.1186/1471-2164-14-446. BMC Genomics. 2013. PMID: 23826801 Free PMC article. - Using Breeding Populations With a Dual Purpose: Cultivar Development and Gene Mapping-A Case Study Using Resistance to Common Bacterial Blight in Dry Bean (Phaseolus vulgaris L.).
Simons KJ, Oladzad A, Lamppa R, Maniruzzaman, McClean PE, Osorno JM, Pasche JS. Simons KJ, et al. Front Plant Sci. 2021 Feb 26;12:621097. doi: 10.3389/fpls.2021.621097. eCollection 2021. Front Plant Sci. 2021. PMID: 33719292 Free PMC article. - Identity by descent: variation in meiosis, across genomes, and in populations.
Thompson EA. Thompson EA. Genetics. 2013 Jun;194(2):301-26. doi: 10.1534/genetics.112.148825. Genetics. 2013. PMID: 23733848 Free PMC article. Review. - Genome-wide analysis reveals adaptation to high altitudes in Tibetan sheep.
Wei C, Wang H, Liu G, Zhao F, Kijas JW, Ma Y, Lu J, Zhang L, Cao J, Wu M, Wang G, Liu R, Liu Z, Zhang S, Liu C, Du L. Wei C, et al. Sci Rep. 2016 May 27;6:26770. doi: 10.1038/srep26770. Sci Rep. 2016. PMID: 27230812 Free PMC article. - Distribution of events of positive selection and population differentiation in a metabolic pathway: the case of asparagine N-glycosylation.
Dall'Olio GM, Laayouni H, Luisi P, Sikora M, Montanucci L, Bertranpetit J. Dall'Olio GM, et al. BMC Evol Biol. 2012 Jun 25;12:98. doi: 10.1186/1471-2148-12-98. BMC Evol Biol. 2012. PMID: 22731960 Free PMC article.
References
Web Resources
- HAP Web site, http://research.calit2.net/hap/
- HaploBlock, http://bioinfo.cs.technion.ac.il/haploblock/
- International HapMap Project, http://www.hapmap.org/
- SeattleSNPs, http://pga.gs.washington.edu
References
- Akaike H (1974) A new look at the statistical model identification. IEEE Trans Automatic Control AC 19:719–723
- Bates JM, Granger CWJ (1969) The combination of forecasts. Oper Res Q 20:451–468
- Breiman L (1996) Bagging predictors. Mach Learn 24:123–140
- Dempster AP, Laird NM, Rubin DB (1977) Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc Ser B 39:1–38
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous