Sleeping Beauty transposase modulates cell-cycle progression through interaction with Miz-1 - PubMed (original) (raw)
Sleeping Beauty transposase modulates cell-cycle progression through interaction with Miz-1
Oliver Walisko et al. Proc Natl Acad Sci U S A. 2006.
Abstract
We used the Sleeping Beauty (SB) transposable element as a tool to probe transposon-host cell interactions in vertebrates. The Miz-1 transcription factor was identified as an interactor of the SB transposase in a yeast two-hybrid screen. Through its association with Miz-1, the SB transposase down-regulates cyclin D1 expression in human cells, as evidenced by differential gene expression analysis using microarray hybridization. Down-regulation of cyclin D1 results in a prolonged G(1) phase of the cell cycle and retarded growth of transposase-expressing cells. G(1) slowdown is associated with a decrease of cyclin D1/cdk4-specific phosphorylation of the retinoblastoma protein. Both cyclin D1 down-regulation and the G(1) slowdown induced by the transposase require Miz-1. A temporary G(1) arrest enhances transposition, suggesting that SB transposition is favored in the G(1) phase of the cell cycle, where the nonhomologous end-joining pathway of DNA repair is preferentially active. Because nonhomologous end-joining is required for efficient SB transposition, the transposase-induced G(1) slowdown is probably a selfish act on the transposon's part to maximize the chance for a successful transposition event.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
Fig. 1.
SB transposase interacts with Miz-1. (A) SB transposase/Miz-1 interaction in a yeast two-hybrid assay. Protein–protein interaction is assessed by activation of the ADE2 and HIS3 reporter genes. (B) In vitro interaction between SB transposase and Miz-1. 35S-labeled Miz-1 was incubated with immobilized maltose binding protein (MBP) or with MBP-SB transposase fusion protein. Bound material recovered after washing was separated by SDS/PAGE and visualized by autoradiography. (C) In vivo interaction between SB transposase and Miz-1. Immunoblot of total extracts of HeLa cells expressing the SB transposase after incubation with antibodies against human Miz-1 or human actin. The blots were hybridized with anti-SB, anti-cyclin D1, and anti-actin antibodies.
Fig. 2.
SB transposase down-regulates cyclin D1 expression. Differential expression of selected cell-cycle regulatory genes in human HeLa cells in response to the presence of the SB transposase. Shown are relative changes of transcript levels in transgenic cells stably expressing the SB transposase compared with wild-type cells, in the presence and absence of a plasmid containing a GFP-tagged transposon (pT/GFP). Cyclin D1 protein levels are reduced in IRES-SB cells, as revealed by Western blot hybridization.
Fig. 3.
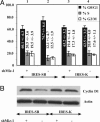
Induction of a G1 slowdown and reduced growth by SB transposase in human HuH7 cells. (A) Growth curves of HuH7 cells expressing SB transposase (IRES-SB) and cells containing the empty vector (IRES-K), established in experiments done in triplicate. The colony-forming assay (six independent platings) shows stained colonies of cells transfected with the empty control (IRES-K), catalytically active SB transposase (IRES-SB), and catalytically inactive SB mutants [IRES-SB (DAE) and IRES-SB (DDD)]. (B) FACScan profiles of IRES-SB and IRES-K cells. Stable expression of SB transposase in IRES-SB cells was verified by Western blot analysis. (C) Levels of phosphorylated retinoblastoma protein in IRES-K and IRES-SB cells were analyzed by Western blot hybridization with antibodies raised against phospho-Rb (Ser-780) and phospho-Rb (Ser 807/811).
Fig. 4.
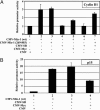
Regulation of the cyclin D1 and p15INK4b promoters by Miz-1, Myc, and SB transposase in transient transfections in HeLa cells. (A) Relative activity of the cyclin D1 promoter in the presence of wild-type and truncated Miz-1 (containing amino acids 269–803), expressed in the presence and absence of Myc and SB transposase. (B) Relative activity of the p15INK4b promoter in the presence of wild-type Miz-1, expressed in the presence and absence of Myc and SB transposase. Data represent fold activation of reporter gene expression. Values obtained in the presence of an empty expression plasmid (CMV) were arbitrarily set to value 1. All experiments were done in triplicate.
Fig. 5.
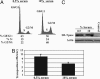
Expression of short hairpin RNA directed against human Miz-1 ablates SB transposase-induced G1 slowdown. (A) Percentages of cells, established in experiments done in triplicate, in different cell-cycle stages of exponentially growing, HuH7-derived IRES-SB, and IRES-K cells, which have either the shMiz-1 construct or the empty vector stably integrated. (B) Cyclin D1 expression levels in IRES-SB and IRES-K cells in the absence and presence of shMiz-1.
Fig. 6.
A temporary G1 arrest by serum-starvation enhances SB transposition in CHO-K1 cells. (A) CHO-K1 cells readily respond to serum withdrawal with a marked G1 arrest. (B) In vivo transposition assay in G1-arrested (serum-starved) and exponentially growing CHO-K1 cells. Transposition efficiency, established in experiments done eight times, represents fold increase in numbers of antibiotic-resistant cell clones in the presence versus in the absence of the transposase. (C) SB transposase expression levels in the presence of 0.5% (serum-starved cells) and 10% serum.
Similar articles
- Induction of G1 cell cycle arrest and P15INK4b expression by ECRG1 through interaction with Miz-1.
Zhao N, Wang J, Cui Y, Guo L, Lu SH. Zhao N, et al. J Cell Biochem. 2004 May 1;92(1):65-76. doi: 10.1002/jcb.20025. J Cell Biochem. 2004. PMID: 15095404 - Sleeping Beauty Transposition.
Ivics Z, Izsvák Z. Ivics Z, et al. Microbiol Spectr. 2015 Apr;3(2):MDNA3-0042-2014. doi: 10.1128/microbiolspec.MDNA3-0042-2014. Microbiol Spectr. 2015. PMID: 26104705 Review. - An alternative pathway for gene regulation by Myc.
Peukert K, Staller P, Schneider A, Carmichael G, Hänel F, Eilers M. Peukert K, et al. EMBO J. 1997 Sep 15;16(18):5672-86. doi: 10.1093/emboj/16.18.5672. EMBO J. 1997. PMID: 9312026 Free PMC article. - Functional zinc finger/sleeping beauty transposase chimeras exhibit attenuated overproduction inhibition.
Wilson MH, Kaminski JM, George AL Jr. Wilson MH, et al. FEBS Lett. 2005 Nov 7;579(27):6205-9. doi: 10.1016/j.febslet.2005.10.004. Epub 2005 Oct 17. FEBS Lett. 2005. PMID: 16243318 - Regulation of cell cycle entry and G1 progression by CSF-1.
Roussel MF. Roussel MF. Mol Reprod Dev. 1997 Jan;46(1):11-8. doi: 10.1002/(SICI)1098-2795(199701)46:1<11::AID-MRD3>3.0.CO;2-U. Mol Reprod Dev. 1997. PMID: 8981358 Review.
Cited by
- RNA-guided retargeting of S_leeping Beauty_ transposition in human cells.
Kovač A, Miskey C, Menzel M, Grueso E, Gogol-Döring A, Ivics Z. Kovač A, et al. Elife. 2020 Mar 6;9:e53868. doi: 10.7554/eLife.53868. Elife. 2020. PMID: 32142408 Free PMC article. - Driving DNA transposition by lentiviral protein transduction.
Cai Y, Mikkelsen JG. Cai Y, et al. Mob Genet Elements. 2014 Jun 23;4:e29591. doi: 10.4161/mge.29591. eCollection 2014. Mob Genet Elements. 2014. PMID: 25057443 Free PMC article. - The piggyBac transposon holds promise for human gene therapy.
Feschotte C. Feschotte C. Proc Natl Acad Sci U S A. 2006 Oct 10;103(41):14981-2. doi: 10.1073/pnas.0607282103. Epub 2006 Oct 2. Proc Natl Acad Sci U S A. 2006. PMID: 17015820 Free PMC article. No abstract available. - The ancient mariner sails again: transposition of the human Hsmar1 element by a reconstructed transposase and activities of the SETMAR protein on transposon ends.
Miskey C, Papp B, Mátés L, Sinzelle L, Keller H, Izsvák Z, Ivics Z. Miskey C, et al. Mol Cell Biol. 2007 Jun;27(12):4589-600. doi: 10.1128/MCB.02027-06. Epub 2007 Apr 2. Mol Cell Biol. 2007. PMID: 17403897 Free PMC article. - Suicidal autointegration of sleeping beauty and piggyBac transposons in eukaryotic cells.
Wang Y, Wang J, Devaraj A, Singh M, Jimenez Orgaz A, Chen JX, Selbach M, Ivics Z, Izsvák Z. Wang Y, et al. PLoS Genet. 2014 Mar 13;10(3):e1004103. doi: 10.1371/journal.pgen.1004103. eCollection 2014 Mar. PLoS Genet. 2014. PMID: 24625543 Free PMC article.
References
- Plasterk R. H., Izsvák Z., Ivics Z. Trends Genet. 1999;15:326–332. - PubMed
- Ivics Z., Hackett P. B., Plasterk R. H., Izsvák Z. Cell. 1997;91:501–510. - PubMed
- Izsvák Z., Ivics Z., Plasterk R. H. J. Mol. Biol. 2000;302:93–102. - PubMed
- Yant S. R., Meuse L., Chiu W., Ivics Z., Izsvák Z., Kay M. A. Nat. Genet. 2000;25:35–41. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials