Restriction of vaccinia virus replication by a ced-3 and ced-4-dependent pathway in Caenorhabditis elegans - PubMed (original) (raw)
Restriction of vaccinia virus replication by a ced-3 and ced-4-dependent pathway in Caenorhabditis elegans
Wan-Hsin Liu et al. Proc Natl Acad Sci U S A. 2006.
Abstract
Genetic tractability and easy manipulation make Caenorhabditis elegans a good model to study host-pathogen interactions. Dozens of different bacterial species can pathogenically infect C. elegans under laboratory conditions, and all of these microbes are extracellular pathogens to nematodes. Viruses, on the other hand, are obligate intracellular parasites, and yet no viral infections have been reported for C. elegans. We established a procedure allowing vaccinia virus to enter and subsequently replicate in C. elegans. Virus replication was significantly enhanced in ced-3, ced-4, ced-9(gf), and egl-1(lf) mutants, demonstrating that the core programmed cell death (PCD) genes ced-3, ced-4, ced-9, and egl-1 control vaccinia virus replication in C. elegans. The ability of ced-3 and ced-4 alleles to restrict virus replication is correlated with their cell-killing activities. Moreover, the increase in vaccinia virus replication levels in the PCD-defective mutants was not likely to be caused by the extra live cells, as neither the inhibition of PCD by icd-1 overexpression nor the presence of extra cells after extra cell divisions in cul-1 or lin-23 mutants had any significant effect on vaccinia virus replication. Therefore, the core PCD genes possess a unique function in controlling vaccinia virus replication in C. elegans.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
Fig. 1.
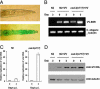
Replication of VV in PEG-treated C. elegans. Groups of about 1000 worms at L3-L4 stage were subjected to PEG-mediated infection by 1 × 107 pfu/ml of vaccinia virus (VV). (A) Wild-type worms exposed to 2% PEG alone (Upper) or to 2% PEG plus 1 × 107 pfu/ml VV (Lower) on the second day postinfection (p.i.) were fixed and stained with X-Gal. (B) PCR analysis. Accumulation of VV genomic DNA was analyzed by PCR in wild-type N2 and ced-3(n717) mutant worms at day 0 or day 3 p.i. (C) Real-time PCR analysis. The relative replication level of VV genomic DNA in wild-type N2 or ced-3(n717) mutant worms was determined by real-time PCR using primers specific for VV B5R gene. The result represents the abundance of replicative DNA in the sample collected at day 3 relative to that at day 0 p.i. The Ct values are shown separately in Table 2, which is published as supporting information on the PNAS web site. All experiments were repeated three times independently and data are means ± SD. (D) Immunoblot. The amounts of VV proteins expressed in N2 and ced-3(n717) mutant at the indicated time points were analyzed by immunoblotting using antibody specific for VV structural D8L protein. For simplicity, we defined day 0 as the time when the 5-min virus exposure with PEG followed by the 6-h virus treatment without PEG had been completed.
Fig. 2.
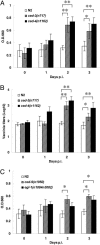
The PCD mutations enhance VV replication in PEG-treated C. elegans. About 1,000 worms (stages L3-L4) of each strain were infected with vaccinia virus in the presence of PEG. (A) VV titers in lysates of ced-3(n717) and ced-4(n1162) mutant strains were measured for β-galactosidase activity at OD660 at different days p.i. (B) After infection, the titers of VV from each C. elegans strain were determined at the indicated days by the standard plaque-forming assay. (C) Virus replication levels in the ced-9(n1950) and egl-1(n1084n3082) mutant strains were determined from the spectrophotometric β-galactosidase activity at OD660 at the indicated times. All experiments were repeated three times independently (∗, P < 0.05; ∗∗, P < 0.01 by Student’s t test). Data are means ± SD.
Fig. 3.
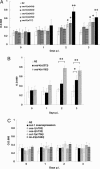
The effects of different ced-3 and ced-4 alleles on vaccinia virus replication in C. elegans. About 1,000 worms (stages L3–L4) of the indicated strains were infected with VV in the presence of PEG. The level of virus replication in each strain was determined from the β-galactosidase activity at OD660. (A) Viral replication levels in most of the ced-3 mutants tested were higher than those in wild type. (B) VV replication levels in ced-4 mutants were significantly higher than those in wild-type worms at the second day p.i. (C) Overexpression of icd-1 or mutations in ces-1, ces-2, cul-1, and lin-23 did not affect VV titers. All experiments were repeated three times. (∗, P < 0.05; ∗∗, P < 0.01 by Student’s t test). Data are means ± SD.
Fig. 4.
Vaccinia-mediated killing of C. elegans is independent of ced-3 and ced-4. About 100 worms (stages L3–L4) of each C. elegans strain were treated under the indicated conditions. Dead worms were picked on indicated days and tested by using X-Gal staining. The percentage of the dead infected worms of each strain was calculated by the following formula: (the number of total X-Gal-positive worms that died by the indicated days/the number of total infected worms) × 100%. The number of total infected worms was calculated by the number of total animals times the infection rate. (A) No PEG or virus treatment. (B) PEG treatment without virus. (C) PEG plus virus treatment. All data (means ± SD) were obtained from four independent experiments.
Comment in
- Worming into the cell: viral reproduction in Caenorhabditis elegans.
Shaham S. Shaham S. Proc Natl Acad Sci U S A. 2006 Mar 14;103(11):3955-6. doi: 10.1073/pnas.0600779103. Epub 2006 Mar 6. Proc Natl Acad Sci U S A. 2006. PMID: 16537467 Free PMC article. No abstract available.
Similar articles
- Phagocytosis promotes programmed cell death in C. elegans.
Reddien PW, Cameron S, Horvitz HR. Reddien PW, et al. Nature. 2001 Jul 12;412(6843):198-202. doi: 10.1038/35084096. Nature. 2001. PMID: 11449278 - The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9.
Conradt B, Horvitz HR. Conradt B, et al. Cell. 1998 May 15;93(4):519-29. doi: 10.1016/s0092-8674(00)81182-4. Cell. 1998. PMID: 9604928 - Adenine nucleotide translocator cooperates with core cell death machinery to promote apoptosis in Caenorhabditis elegans.
Shen Q, Qin F, Gao Z, Cui J, Xiao H, Xu Z, Yang C. Shen Q, et al. Mol Cell Biol. 2009 Jul;29(14):3881-93. doi: 10.1128/MCB.01509-08. Epub 2009 May 4. Mol Cell Biol. 2009. PMID: 19414600 Free PMC article. - Programmed cell death in Caenorhabditis elegans.
Hengartner MO, Horvitz HR. Hengartner MO, et al. Curr Opin Genet Dev. 1994 Aug;4(4):581-6. doi: 10.1016/0959-437x(94)90076-f. Curr Opin Genet Dev. 1994. PMID: 7950327 Review. - egl-1: a key activator of apoptotic cell death in C. elegans.
Nehme R, Conradt B. Nehme R, et al. Oncogene. 2008 Dec;27 Suppl 1:S30-40. doi: 10.1038/onc.2009.41. Oncogene. 2008. PMID: 19641505 Review.
Cited by
- Expression of hepatitis B virus surface antigens induces defective gonad phenotypes in Caenorhabditis elegans.
Chen YY, Lee LW, Hong WN, Lo SJ. Chen YY, et al. World J Virol. 2017 Feb 12;6(1):17-25. doi: 10.5501/wjv.v6.i1.17. World J Virol. 2017. PMID: 28239568 Free PMC article. - The response of mammalian cells to double-stranded RNA.
Gantier MP, Williams BR. Gantier MP, et al. Cytokine Growth Factor Rev. 2007 Oct-Dec;18(5-6):363-71. doi: 10.1016/j.cytogfr.2007.06.016. Epub 2007 Aug 14. Cytokine Growth Factor Rev. 2007. PMID: 17698400 Free PMC article. Review. - Systems Biology of Virus-Host Protein Interactions: From Hypothesis Generation to Mechanisms of Replication and Pathogenesis.
Shah PS, Beesabathuni NS, Fishburn AT, Kenaston MW, Minami SA, Pham OH, Tucker I. Shah PS, et al. Annu Rev Virol. 2022 Sep 29;9(1):397-415. doi: 10.1146/annurev-virology-100520-011851. Epub 2022 May 16. Annu Rev Virol. 2022. PMID: 35576593 Free PMC article. Review. - An evolutionarily conserved transcriptional response to viral infection in Caenorhabditis nematodes.
Chen K, Franz CJ, Jiang H, Jiang Y, Wang D. Chen K, et al. BMC Genomics. 2017 Apr 17;18(1):303. doi: 10.1186/s12864-017-3689-3. BMC Genomics. 2017. PMID: 28415971 Free PMC article. - Transport of sequence-specific RNA interference information between cells.
Jose AM, Hunter CP. Jose AM, et al. Annu Rev Genet. 2007;41:305-30. doi: 10.1146/annurev.genet.41.110306.130216. Annu Rev Genet. 2007. PMID: 17645412 Free PMC article. Review.
References
- Hodgkin J., Kuwabara P. E., Corneliussen B. Curr. Biol. 2000;10:1615–1618. - PubMed
- Kurz C. L., Ewbank J. J. Nat. Rev. Genet. 2003;4:380–390. - PubMed
- Schulenburg H., Kurz C. L., Ewbank J. J. Immunol. Rev. 2004;198:36–58. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources