Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta - PubMed (original) (raw)
Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta
Martin E Budd et al. Mol Cell Biol. 2006 Apr.
Abstract
The precise machineries required for two aspects of eukaryotic DNA replication, Okazaki fragment processing (OFP) and telomere maintenance, are poorly understood. In this work, we present evidence that Saccharomyces cerevisiae Pif1 helicase plays a wider role in DNA replication than previously appreciated and that it likely functions in conjunction with Dna2 helicase/nuclease as a component of the OFP machinery. In addition, we show that Dna2, which is known to associate with telomeres in a cell-cycle-specific manner, may be a new component of the telomere replication apparatus. Specifically, we show that deletion of PIF1 suppresses the lethality of a DNA2-null mutant. The pif1delta dna2delta strain remains methylmethane sulfonate sensitive and temperature sensitive; however, these phenotypes can be suppressed by further deletion of a subunit of pol delta, POL32. Deletion of PIF1 also suppresses the cold-sensitive lethality and hydroxyurea sensitivity of the pol32delta strain. Dna2 is thought to function by cleaving long flaps that arise during OFP due to excessive strand displacement by pol delta and/or by an as yet unidentified helicase. Thus, suppression of dna2delta can be rationalized if deletion of POL32 and/or PIF1 results in a reduction in long flaps that require Dna2 for processing. We further show that deletion of DNA2 suppresses the long-telomere phenotype and the high rate of formation of gross chromosomal rearrangements in pif1Delta mutants, suggesting a role for Dna2 in telomere elongation in the absence of Pif1.
Figures
FIG. 1.
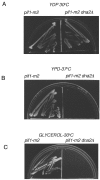
Suppression of dna2 mutant phenotypes by deletion of PIF1. (A) Serial dilutions of identical numbers of cells were carried out at the temperatures indicated. The following strains were used: W303, WT; MB90-7A, dna2-1; MB91, _dna2-1 pif1_Δ. (B) Serial dilutions of identical numbers of each strain were plated on medium containing the indicated amounts of MMS. Relevant genotypes are indicated. The following strains were used: BY4741, WT; 10509, _pif1_Δ; 4741dna2-2, dna2-2; MB208, _dna2-2 pif1_Δ; and MB203, _dna2_Δ _pif1_Δ (Table 1).
FIG. 2.
Suppression of _dna2_Δ lethality by inactivation of Pif1 nuclear function but not by inactivation of Pif1 mitochondrial function. The following strains were used: 4344, pif1-m2; and 4344dna2Δ, _pif1-m2 dna2_Δ. (A and B) Growth on glucose-containing medium. (C) Growth on nonfermentable glycerol medium.
FIG. 3.
Reduced DNA damage in _dna2-1 pif1_Δ. The strains used are isogenic derivatives of strain W303—MB90-7A, MB91, U953-61, MB92-M, SPY40, MB92-31C, MB92-35A, and U960-5C (Table 1)—with one exception, the _pif1_Δ strain MB509. The relevant genotypes are indicated in the figure. Strains were grown to approximately 1 × 107 per ml. Extracts were prepared and Western blots performed using antibody against Rad53 (gift of John Diffley, Clare Hall, England) as described previously (16). Wild-type W303 was also treated with 0.02% MMS where indicated to show phosphorylated Rad53. (A) Rad53 is phosphorylated in dna2-1 and dna2-2 strains at 23°C in a _MEC1_-dependent and _TEL1_-independent fashion. The 100-kDa marker runs with fully phosphorylated Rad53 and is indicated by the arrow in the figure. The asterisk denotes a nonspecific, cross-reacting species. (B) Rad53 is phosphorylated in dna2-1 and dna2-2 strains at 37°C in a _MEC1_-dependent and _TEL1_-independent manner. (C) _pif1_Δ suppresses Rad53 phosphorylation in the dna2-1 strain.
FIG. 4.
Suppression of the temperature sensitivity of the _dna2_Δ _pif1_Δ strain by _pol32_Δ. The following strains were incubated on a YPD plate at 37°C for 3 days: MB202, _pif1_Δ; MB203.6, _dna2_Δ _pif1_Δ; MB206, _pif1_Δ _pol32_Δ; and MB207, _dna2_Δ _pif1_Δ _pol32_Δ.
FIG. 5.
Suppression of the cold sensitivity and HU sensitivity of the _pol32_Δ strain by _pif1_Δ. The following BY4741 isogenic strains were used: BY4741, WT; MB205, _pol32_Δ; MB201, _pif1_Δ; and MB206, _pol32_Δ _pif1_Δ. YPD plates were spotted with these strains and incubated at 30°C for 2 days (top, left panel) or at 16°C for 7 days (top right panel) as indicated. YPD plates were spotted with the same strains in the absence (lower left) or presence (lower right) of 38 mM HU and incubated at 23°C.
FIG. 6.
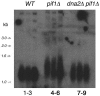
Deletion of DNA2 reduces telomere length in _pif1_Δ strains. Lanes 1, 2, and 3, WT strains 47421, BY4741, and BY4742, respectively. Lanes 4, 5, and 6, three colonies of _pif1_Δ strain 10509. Lanes 7, 8, and 9, _dna2_Δ _pif1_Δ strains MB203.3, MB203.5, and MB203.6, respectively. MB203.3, MB203.5, and MB203.6 are three independent _pif1_Δ transformants of MB110.
FIG. 7.
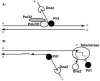
Interpretation of genetic interactions between PIF1 and DNA2. (A) Pif1 may help to make a flap during RNA removal by Dna2 during OFP. (B) Dna2 may be required for optimum elongation of telomeres by telomerase, whereas Pif1 has been shown to inhibit telomerase processivity (5). Pif1 may have a second role at telomeres in which it aids Dna2 in removal of RNA from the Okazaki fragments at the telomere. The last Okazaki fragment might have a special requirement for Pif1 helicase as there is no pol δ/PCNA present to strand displace and to recruit FEN1.
Similar articles
- Flexibility of eukaryotic Okazaki fragment maturation through regulated strand displacement synthesis.
Stith CM, Sterling J, Resnick MA, Gordenin DA, Burgers PM. Stith CM, et al. J Biol Chem. 2008 Dec 5;283(49):34129-40. doi: 10.1074/jbc.M806668200. Epub 2008 Oct 16. J Biol Chem. 2008. PMID: 18927077 Free PMC article. - Inviability of a DNA2 deletion mutant is due to the DNA damage checkpoint.
Budd ME, Antoshechkin IA, Reis C, Wold BJ, Campbell JL. Budd ME, et al. Cell Cycle. 2011 May 15;10(10):1690-8. doi: 10.4161/cc.10.10.15643. Epub 2011 May 15. Cell Cycle. 2011. PMID: 21508669 Free PMC article. - Disease-associated DNA2 nuclease-helicase protects cells from lethal chromosome under-replication.
Falquet B, Ölmezer G, Enkner F, Klein D, Challa K, Appanah R, Gasser SM, Rass U. Falquet B, et al. Nucleic Acids Res. 2020 Jul 27;48(13):7265-7278. doi: 10.1093/nar/gkaa524. Nucleic Acids Res. 2020. PMID: 32544229 Free PMC article. - Yet another job for Dna2: Checkpoint activation.
Wanrooij PH, Burgers PM. Wanrooij PH, et al. DNA Repair (Amst). 2015 Aug;32:17-23. doi: 10.1016/j.dnarep.2015.04.009. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 25956863 Free PMC article. Review. - DNA2-An Important Player in DNA Damage Response or Just Another DNA Maintenance Protein?
Pawłowska E, Szczepanska J, Blasiak J. Pawłowska E, et al. Int J Mol Sci. 2017 Jul 18;18(7):1562. doi: 10.3390/ijms18071562. Int J Mol Sci. 2017. PMID: 28718810 Free PMC article. Review.
Cited by
- Mammalian Resilience Revealed by a Comparison of Human Diseases and Mouse Models Associated With DNA Helicase Deficiencies.
Kohzaki M. Kohzaki M. Front Mol Biosci. 2022 Aug 11;9:934042. doi: 10.3389/fmolb.2022.934042. eCollection 2022. Front Mol Biosci. 2022. PMID: 36032672 Free PMC article. Review. - The secondary-structured DNA-binding activity of Dna2 endonuclease/helicase is critical to cell growth under replication stress.
Park S, Karatayeva N, Demin AA, Munashingha PR, Seo YS. Park S, et al. FEBS J. 2021 Feb;288(4):1224-1242. doi: 10.1111/febs.15475. Epub 2020 Jul 19. FEBS J. 2021. PMID: 32638513 Free PMC article. - Chromosome Duplication in Saccharomyces cerevisiae.
Bell SP, Labib K. Bell SP, et al. Genetics. 2016 Jul;203(3):1027-67. doi: 10.1534/genetics.115.186452. Genetics. 2016. PMID: 27384026 Free PMC article. Review. - Mammalian DNA2 helicase/nuclease cleaves G-quadruplex DNA and is required for telomere integrity.
Lin W, Sampathi S, Dai H, Liu C, Zhou M, Hu J, Huang Q, Campbell J, Shin-Ya K, Zheng L, Chai W, Shen B. Lin W, et al. EMBO J. 2013 May 15;32(10):1425-39. doi: 10.1038/emboj.2013.88. Epub 2013 Apr 19. EMBO J. 2013. PMID: 23604072 Free PMC article. - Telomerase-null survivor screening identifies novel telomere recombination regulators.
Hu Y, Tang HB, Liu NN, Tong XJ, Dang W, Duan YM, Fu XH, Zhang Y, Peng J, Meng FL, Zhou JQ. Hu Y, et al. PLoS Genet. 2013;9(1):e1003208. doi: 10.1371/journal.pgen.1003208. Epub 2013 Jan 17. PLoS Genet. 2013. PMID: 23390378 Free PMC article.
References
- Ayyagari, R., X. V. Gomes, D. A. Gordenin, and P. M. J. Burgers. 2003. Okazaki fragment maturation in yeast. I. Distribution of functions between Fen1 and Dna2. J. Biol. Chem. 278:1618-1625. - PubMed
- Bae, S. H., K.-H. Bae, J. A. Kim, and Y. S. Seo. 2001. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412:456-461. - PubMed
- Bessler, J. B., J. Z. Torre, and V. A. Zakian. 2001. The Pif1p subfamily of helicases: region-specific DNA helicases? Trends Cell Biol. 11:60. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases