Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin - PubMed (original) (raw)
Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin
Emily Bernstein et al. Mol Cell Biol. 2006 Apr.
Abstract
The chromodomain (CD) of the Drosophila Polycomb protein exhibits preferential binding affinity for histone H3 when trimethylated at lysine 27. Here we have investigated the five mouse Polycomb homologs known as Cbx2, Cbx4, Cbx6, Cbx7, and Cbx8. Despite a high degree of conservation, the Cbx chromodomains display significant differences in binding preferences. Not all CDs bind preferentially to K27me3; rather, some display affinity towards both histone H3 trimethylated at K9 and H3K27me3, and one CD prefers K9me3. Cbx7, in particular, displays strong affinity for both H3K9me3 and H3K27me3 and is developmentally regulated in its association with chromatin. Cbx7 associates with facultative heterochromatin and, more specifically, is enriched on the inactive X chromosome. Finally, we find that, in vitro, the chromodomain of Cbx7 can bind RNA and that, in vivo, the interaction of Cbx7 with chromatin, and the inactive X chromosome in particular, depends partly on its association with RNA. We propose that the capacity of this mouse Polycomb homolog to associate with the inactive X chromosome, or any other region of chromatin, depends not only on its chromodomain but also on the combination of histone modifications and RNA molecules present at its target sites.
Figures
FIG. 1.
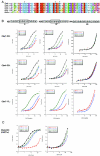
Analysis of Pc-like Cbx CD binding affinities for trimethylated H3K9 and H3K27. (A) ClustalW alignment of the five mouse Pc-like CDs (aa 1 to 62) and the Drosophila Pc CD (aa 16 to 78). The asterisks represent the caging aromatic residues that mediate the histone methyl-lysine interaction. Note the high degree of conservation among these family members. (B) Fluorescence polarization of Cbx7 (top) and Cbx4 (middle) to histone tail peptides, including the me1, me2, and me3 states on residues K9 and K27 of H3, and K20me1, K20me2, and K20me3 of H4, respectively. Full-length Cbx7 (bottom) was tested against all of the above except for the monomethylated forms of H3K9 and H3K27, H4K20me2 and H4K20me3, and behaves identically to its CD for peptides tested. Histone tail sequences are represented above; note the ARKS motifs of H3K9 and H3K27. See panel D for actual peptides used. (C) Fluorescence polarization of Cbx5 (mouse HP1α) to H3K9 and H3K27 histone tail peptides in the me1, me2, and me3 states. (D) Dissociation constants (Kd, in micromolars) for Cbx2, Cbx4, Cbx6, Cbx7, and Cbx8, as well as Cbx5 (mHP1α), with each series of methylated peptides for each backbone shown. Low-micromolar binding constants are highlighted in red. The asterisk depicts weak binding of Cbx7 for H3K9me2 (a mark that is enriched on the Xi). Values represent averages ± standard deviations for at least three independent experiments in all cases (except for Cbx5 with certain peptides). ND, not determined. (E) Peptide pull-down assays. (Left) All CDs were examined for binding to unmodified and trimethylated peptides representing H3K9, H3K27, and H4K20. Results support those obtained by FP (D). (Right) Cbx7 CD, Cbx7 caging aromatic point mutant F11A, and human HP1β CD recombinant proteins were tested for the ability to bind unmodified and me1, me2, and me3 peptides of H3K9 and K27. Note the trimethyl specificity of Cbx7. (Right, bottom) GST and full-length Cbx7 were examined for binding to unmodified and trimethylated biotinylated peptides of H3K9 and H3K27. Full-length Cbx7 behaves the same as its CD alone. GST does not bind any peptide, as expected. un, unmodified.
FIG. 1.
Analysis of Pc-like Cbx CD binding affinities for trimethylated H3K9 and H3K27. (A) ClustalW alignment of the five mouse Pc-like CDs (aa 1 to 62) and the Drosophila Pc CD (aa 16 to 78). The asterisks represent the caging aromatic residues that mediate the histone methyl-lysine interaction. Note the high degree of conservation among these family members. (B) Fluorescence polarization of Cbx7 (top) and Cbx4 (middle) to histone tail peptides, including the me1, me2, and me3 states on residues K9 and K27 of H3, and K20me1, K20me2, and K20me3 of H4, respectively. Full-length Cbx7 (bottom) was tested against all of the above except for the monomethylated forms of H3K9 and H3K27, H4K20me2 and H4K20me3, and behaves identically to its CD for peptides tested. Histone tail sequences are represented above; note the ARKS motifs of H3K9 and H3K27. See panel D for actual peptides used. (C) Fluorescence polarization of Cbx5 (mouse HP1α) to H3K9 and H3K27 histone tail peptides in the me1, me2, and me3 states. (D) Dissociation constants (Kd, in micromolars) for Cbx2, Cbx4, Cbx6, Cbx7, and Cbx8, as well as Cbx5 (mHP1α), with each series of methylated peptides for each backbone shown. Low-micromolar binding constants are highlighted in red. The asterisk depicts weak binding of Cbx7 for H3K9me2 (a mark that is enriched on the Xi). Values represent averages ± standard deviations for at least three independent experiments in all cases (except for Cbx5 with certain peptides). ND, not determined. (E) Peptide pull-down assays. (Left) All CDs were examined for binding to unmodified and trimethylated peptides representing H3K9, H3K27, and H4K20. Results support those obtained by FP (D). (Right) Cbx7 CD, Cbx7 caging aromatic point mutant F11A, and human HP1β CD recombinant proteins were tested for the ability to bind unmodified and me1, me2, and me3 peptides of H3K9 and K27. Note the trimethyl specificity of Cbx7. (Right, bottom) GST and full-length Cbx7 were examined for binding to unmodified and trimethylated biotinylated peptides of H3K9 and H3K27. Full-length Cbx7 behaves the same as its CD alone. GST does not bind any peptide, as expected. un, unmodified.
FIG. 2.
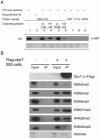
Cbx7-associated histone modifications. (A) Far Western blotting assay. The GST-Cbx7 CD binds to female ES cell (LF2) histone H3 through K9me3 and K27me3, demonstrated by peptide competitions (lanes 1 to 6); Cbx7 CD does not bind to recombinant H3 (lane 2); GST alone and caging aromatic mutants fail to associate (lanes 7 to 9); Ponceau for equal loading of total histones (bottom). (B) IPs of mononucleosomal extracts prepared from Cbx7-flag stable 293 cells versus control cells. Inputs in both cases show presence of all histone modifications; Cbx7-flag associated histones (IP) are marked predominantly by facultative heterochromatic modifications, including H3K9me2, H3K27me3, and H4K20me1. un, unmodified.
FIG. 3.
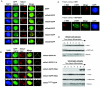
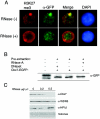
Cbx proteins associate with the inactive X. (A) All Cbx-EGFP fusion proteins, except Cbx4, localize to the Xi in 3-day-differentiated female ES cells. The Xi is visualized by K27me3 staining (red). Fluorescence intensity plots (white line from a to b) across the nucleus and including the K27me3 signal (red) on the Xi illustrate the enrichment for, or lack of, each of the Cbx-GFP fusion proteins (green). (B) Point mutations of the caging aromatic residues in the Cbx7 CD (F11A, W32A, and W35A) disrupt localization to the Xi. When the CD of Cbx7 is replaced by the CD of Cbx4 (mCbx7Cbx4CD-EGFP), the chimeric protein no longer strongly associates with the Xi, although it is not disrupted completely (bottom panel). Note that the K27me3 peak coincides with a Cbx7-GFP peak for the wild-type (WT) and Cbx7Cbx4CD proteins but not for the three mutated versions. (C) GST overlay assays were performed on 3-day-differentiated female mouse ES cells. Protein was detected on paraformaldehyde-fixed cells using α-GST antibody (green). The Xi was visualized by α-Eed antibody (red). GST protein does not associate with the Xi (top), while full-length GST-Cbx7 is enriched on the Xi (bottom). It should be noted that although these experiments support our EGFP-fusion results, they were somewhat variable in their efficiency. (D) Female ES cells were tested throughout differentiation (up to day 7) for Cbx7 association with chromatin (un, undifferentiated). Cbx7 and K27me3 are expressed throughout the differentiation process (top, whole-cell extracts); however, Cbx7 specifically associates with chromatin on day 6 and onward (compare to HP1β and K27me3 in chromatin extracts at the bottom of the panel).
FIG. 4.
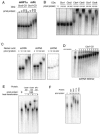
Pc-like Cbx CDs bind RNA. Recombinant CDs were tested for RNA binding by gel shift assays. (A) The Cbx5 (mouse HP1α) CD was compared to Cbx7 (mPc) for RNA binding with a 500-nt single stranded RNA (ssRNA), and only the latter demonstrates a shift. (B) Cbx4, Cbx6, Cbx7, and Cbx8 CDs interact with RNA, while Cbx1 (mHP1β) and Cbx2 do not. (C) Cbx7 binds to both single-stranded RNA (ssRNA) and double-stranded RNA (dsRNA). Cbx7 binds dsDNA with only minor affinity. (D) Gel shift analysis of the Cbx7 CD with ssRNA (500mer) was performed by extensive titration of protein in order to determine a dissociation constant, which is approximately 100 μM. (E) Heat denaturation of Cbx6, Cbx8, and dPc CD proteins disrupts the interaction with RNA (+, heat denaturation). (F) Point mutations in the caging aromatic residues of the Cbx7 CD do not abrogate RNA binding. Recombinant GST was used as a negative control.
FIG. 5.
Cbx7 chromatin association is RNA dependent. (A) RNase treatment of 3-day-differentiated ES cells strongly diminished the accumulation of Cbx7-EGFP on the Xi. The Xi is visualized by K27me3 staining (red), and the signal of Cbx7-EGFP was enhanced using α-GFP antibody (green). (B) Degradation of Cbx7 is not observed after RNase treatment (or DNase I treatment), as detected by Western blot with GFP antibody. (C) Cbx7 is depleted from 6-day-differentiated ES cell chromatin by RNase treatment (top), while WDR5 and HP1β are not affected. See histones for equal loading (bottom).
Similar articles
- Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27.
Min J, Zhang Y, Xu RM. Min J, et al. Genes Dev. 2003 Aug 1;17(15):1823-8. doi: 10.1101/gad.269603. Genes Dev. 2003. PMID: 12897052 Free PMC article. - Different polycomb group CBX family proteins associate with distinct regions of chromatin using nonhomologous protein sequences.
Vincenz C, Kerppola TK. Vincenz C, et al. Proc Natl Acad Sci U S A. 2008 Oct 28;105(43):16572-7. doi: 10.1073/pnas.0805317105. Epub 2008 Oct 16. Proc Natl Acad Sci U S A. 2008. PMID: 18927235 Free PMC article. - Interaction proteomics analysis of polycomb proteins defines distinct PRC1 complexes in mammalian cells.
Vandamme J, Völkel P, Rosnoblet C, Le Faou P, Angrand PO. Vandamme J, et al. Mol Cell Proteomics. 2011 Apr;10(4):M110.002642. doi: 10.1074/mcp.M110.002642. Epub 2011 Jan 31. Mol Cell Proteomics. 2011. PMID: 21282530 Free PMC article. - Biological functions of chromobox (CBX) proteins in stem cell self-renewal, lineage-commitment, cancer and development.
van Wijnen AJ, Bagheri L, Badreldin AA, Larson AN, Dudakovic A, Thaler R, Paradise CR, Wu Z. van Wijnen AJ, et al. Bone. 2021 Feb;143:115659. doi: 10.1016/j.bone.2020.115659. Epub 2020 Sep 24. Bone. 2021. PMID: 32979540 Review. - Phosphorylation of repressive histone code readers by casein kinase 2 plays diverse roles in heterochromatin regulation.
Murakami Y. Murakami Y. J Biochem. 2019 Jul 1;166(1):3-6. doi: 10.1093/jb/mvz045. J Biochem. 2019. PMID: 31198932 Review.
Cited by
- Polycomb and trithorax opposition in development and disease.
Poynter ST, Kadoch C. Poynter ST, et al. Wiley Interdiscip Rev Dev Biol. 2016 Nov;5(6):659-688. doi: 10.1002/wdev.244. Epub 2016 Sep 1. Wiley Interdiscip Rev Dev Biol. 2016. PMID: 27581385 Free PMC article. Review. - RYBP stimulates PRC1 to shape chromatin-based communication between Polycomb repressive complexes.
Rose NR, King HW, Blackledge NP, Fursova NA, Ember KJ, Fischer R, Kessler BM, Klose RJ. Rose NR, et al. Elife. 2016 Oct 5;5:e18591. doi: 10.7554/eLife.18591. Elife. 2016. PMID: 27705745 Free PMC article. - Epigenetic modifiers: activities in renal cell carcinoma.
de Cubas AA, Rathmell WK. de Cubas AA, et al. Nat Rev Urol. 2018 Oct;15(10):599-614. doi: 10.1038/s41585-018-0052-7. Nat Rev Urol. 2018. PMID: 30030490 Free PMC article. Review. - Functional Landscape of PCGF Proteins Reveals Both RING1A/B-Dependent-and RING1A/B-Independent-Specific Activities.
Scelfo A, Fernández-Pérez D, Tamburri S, Zanotti M, Lavarone E, Soldi M, Bonaldi T, Ferrari KJ, Pasini D. Scelfo A, et al. Mol Cell. 2019 Jun 6;74(5):1037-1052.e7. doi: 10.1016/j.molcel.2019.04.002. Epub 2019 Apr 24. Mol Cell. 2019. PMID: 31029542 Free PMC article. - Acquisition of neural fate by combination of BMP blockade and chromatin modification.
Ong ALC, Kokaji T, Kishi A, Takihara Y, Shinozuka T, Shimamoto R, Isotani A, Shirai M, Sasai N. Ong ALC, et al. iScience. 2023 Sep 9;26(10):107887. doi: 10.1016/j.isci.2023.107887. eCollection 2023 Oct 20. iScience. 2023. PMID: 37771660 Free PMC article.
References
- Akhtar, A., and P. B. Becker. 2000. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell 5:367-375. - PubMed
- Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. - PubMed
- Bernstein, E., and C. D. Allis. 2005. RNA meets chromatin. Genes Dev. 19:1635-1655. - PubMed
- Boggs, B. A., P. Cheung, E. Heard, D. L. Spector, A. C. Chinault, and C. D. Allis. 2002. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. 30:73-76. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases