cis- and trans-Acting determinants of transcription termination by yeast RNA polymerase II - PubMed (original) (raw)
cis- and trans-Acting determinants of transcription termination by yeast RNA polymerase II
Eric J Steinmetz et al. Mol Cell Biol. 2006 Apr.
Abstract
Most eukaryotic genes are transcribed by RNA polymerase II (Pol II), including those that produce mRNAs and many noncoding functional RNAs. Proper expression of these genes requires efficient termination by Pol II to avoid transcriptional interference and synthesis of extended, nonfunctional RNAs. We previously described a pathway for yeast Pol II termination that involves recognition of an element in the nascent transcript by the essential RNA-binding protein Nrd1. The Nrd1-dependent pathway appears to be used primarily for nonpolyadenylated transcripts, such as the small nuclear and small nucleolar RNAs (snoRNAs). mRNAs are thought to use a distinct pathway that is coupled to cleavage and polyadenylation of the transcript. Here we show that the terminator elements for two yeast snoRNA genes also direct polyadenylated 3'-end formation in the context of an mRNA 3' untranslated region. A selection for cis-acting terminator readthrough mutations identified conserved features of these elements, some of which are similar to cleavage and polyadenylation signals. A selection for trans-acting mutations that induce readthrough of both a snoRNA and an mRNA terminator yielded mutations in the Rpb3 and Rpb11 subunits of Pol II that define a remarkably discrete surface on the trailing end of the enzyme. Our results suggest that, at least in budding yeast, protein-coding and noncoding Pol II-transcribed genes use similar mechanisms to direct termination and that the termination signal is transduced through the Rpb3/Rpb11 heterodimer.
Figures
FIG. 1.
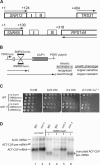
(A) Locations of terminator elements downstream of the SNR13 and SNR65 snoRNA genes. Numbers indicate position in base pairs from the transcription start site of the snoRNA genes. Boxes with heavy outlines indicate either the coding sequence for the mature snoRNAs snR13 and snR65 or the open reading frames for the downstream protein-coding genes. Arrows indicate the directions of transcription. Boxes with thin outlines demarcate the region I and region II terminator elements of SNR13 (39) or SNR65 (this study). (B) Scheme for selection of _cis_-acting mutations in snoRNA terminators (term.). See text for details. (C) Copper plate assay for SNR65 terminator function. Tenfold serial dilutions of cultures of the 46α _cup1_Δ (WT [wild type]) and _nrd1_-5 mutant strains harboring ACT/CUP reporter plasmids with no insert or with the indicated SNR65 3′-flanking sequences (SNR65long, bp 101 to 317; SNR65short, bp 101 to 236) inserted into the intron were spotted onto medium lacking leucine and containing the indicated concentrations of copper and incubated for 2 to 3 days at 30°C. (D) Northern analysis of ACT/CUP RNA. Total cellular RNA was prepared from the 46α (WT) and _nrd1_-5 mutant strains harboring ACT/CUP reporter plasmids with no insert or with SNR65long (L) or SNR65short (S) sequences inserted into the intron. The blot was probed with 32P-labeled oligonucleotide ACT1-probe, which is complementary to the ACT1 mRNA 5′ UTR and so detects ACT/CUP mRNA, as well as endogenous ACT1 mRNA.
FIG. 2.
Termination-polyadenylation elements of snR13 and snR65 pre-snoRNAs. Nucleotide positions relative to the transcription start site and mature 5′ end are shown above and below the sequences for snR13 and snR65, respectively. Lowercase letters indicate nucleotides contained within mature snR13, while uppercase letters indicate nucleotides downstream of the mature 3′ end of either RNA. Also shown is a portion of the NRD1 5′ UTR (nucleotides −229 to −173 relative to the start codon) that contains a Nrd1-responsive element (40). Sequences that are identical in two or more of the genes are in boldface. Residues at which mutations decrease terminator function in the context of the ACT/CUP fusion gene intron, either singly or in combination (see Table 1), are boxed. SNR13 and SNR65 polyadenylation sites in the context of the ACT/CUP fusion gene 3′ UTR are indicated by filled stars (major sites) or open stars (additional sites).
FIG. 3.
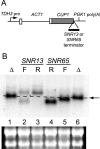
(A) Scheme for mapping of functional polyadenylation sites in snoRNA terminators. The CUP1 3′ UTR (157 nucleotides) was deleted prior to insertion of either the SNR13 or the SNR65 terminator at the site of the deletion. (B) Northern analysis of transcripts from ACT/CUP fusion gene alleles. Total cellular RNA was extracted from the 46α _cup1_Δ strain carrying plasmid-borne ACT/CUP alleles with the following modifications: lanes 1 and 6, deletion of 157 nucleotides of the CUP1 3′ UTR (Δ); lanes 2 and 3, insertion of SNR13 nucleotides +125 to +232 in the forward (F; lane 2) and reverse (R; lane 3) orientations; lanes 4 and 5, insertion of SNR65 nucleotides +101 to +317 in the reverse (lane 4) and forward (lane 5) orientations. The arrow and dashed line mark the position of mRNA with only the CUP1 3′ UTR deletion. The blot was probed with 32P-labeled oligonucleotide 3′CUP-2, complementary to the CUP1 coding region. Ethidium bromide staining of the portion of the filter containing the large and small rRNAs is shown below the lane numbers, for normalization.
FIG. 4.
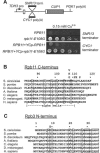
Terminator readthrough mutations in the Rpb11 and Rpb3 subunits of Pol II. (A) Scheme for selecting _trans_-acting mutations that induce readthrough of either the snR13 snoRNA gene terminator or the Cyc1 protein-coding gene terminator. In the examples shown, the _rpb11_-E108G mutation induces readthrough of the SNR13 terminator when present in the genome and dominantly induces readthrough of the CYC1 terminator when present on a low-copy-number plasmid. (B) Readthrough substitutions isolated in Rpb11 are located at the C terminus and are shown above an alignment of Rpb11 orthologs. Only single-residue substitutions are shown (see Table 2). Numbers refer only to residues in the S. cerevisiae protein. Conserved hydrophobic residues in the α-helical segment are boxed. Humans have several isoforms of Rpb11 that differ at the C terminus (21); isoform a is shown. (C) The readthrough substitution isolated in Rpb3 is located at the N terminus and is shown above an alignment of Rpb3 orthologs. The most-conserved residues are boxed.
FIG. 5.
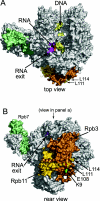
Shown are top (A) and rear (B) orthogonal views of 12-subunit yeast Pol II with a transcription bubble mimic in the active site (27). DNA (pale yellow) enters from the top in panel A and exits toward the viewer. RNA (orange) exits through a pore that is covered in part by the lid (magenta). Rpb3 (copper) and Rpb11 (gold) are at the back of Pol II, while Rpb7 (pale green) extends from the side. Rpb11 residue E108 is red, while L111 and L114 are gold. (Residues 115 to 120 are not resolved in the crystal structure.) Rpb3 residue K9 is bright green. Rpb11-E108 is close to Rpb3-K9 in the back view. The C termini of Rpb3 and Rpb11 are shown as ribbons to emphasize the coiled-coil structure. This figure was created with PyMOL (DeLano Scientific) by using pdb file 1Y1W (
).
Similar articles
- Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription.
Steinmetz EJ, Brow DA. Steinmetz EJ, et al. Mol Cell Biol. 2003 Sep;23(18):6339-49. doi: 10.1128/MCB.23.18.6339-6349.2003. Mol Cell Biol. 2003. PMID: 12944462 Free PMC article. - Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts.
Carroll KL, Pradhan DA, Granek JA, Clarke ND, Corden JL. Carroll KL, et al. Mol Cell Biol. 2004 Jul;24(14):6241-52. doi: 10.1128/MCB.24.14.6241-6252.2004. Mol Cell Biol. 2004. PMID: 15226427 Free PMC article. - RNA-binding protein Nrd1 directs poly(A)-independent 3'-end formation of RNA polymerase II transcripts.
Steinmetz EJ, Conrad NK, Brow DA, Corden JL. Steinmetz EJ, et al. Nature. 2001 Sep 20;413(6853):327-31. doi: 10.1038/35095090. Nature. 2001. PMID: 11565036 - [Molecular mechanisms of coupling processes of transcription termination and polyadenylation of pro-mRNA].
Zarudnaia MI, Potiagaĭ AL, Dzerzhinskiĭ NE, Gororun DN. Zarudnaia MI, et al. Ukr Biokhim Zh (1999). 2001 Mar-Apr;73(2):28-32. Ukr Biokhim Zh (1999). 2001. PMID: 11642040 Review. Russian. - Implications of polyadenylation in health and disease.
Curinha A, Oliveira Braz S, Pereira-Castro I, Cruz A, Moreira A. Curinha A, et al. Nucleus. 2014;5(6):508-19. doi: 10.4161/nucl.36360. Epub 2014 Oct 31. Nucleus. 2014. PMID: 25484187 Free PMC article. Review.
Cited by
- The exosome component Rrp6 is required for RNA polymerase II termination at specific targets of the Nrd1-Nab3 pathway.
Fox MJ, Gao H, Smith-Kinnaman WR, Liu Y, Mosley AL. Fox MJ, et al. PLoS Genet. 2015 Feb 13;11(2):e1004999. doi: 10.1371/journal.pgen.1004999. eCollection 2015. PLoS Genet. 2015. PMID: 25680078 Free PMC article. - Fail-safe transcriptional termination for protein-coding genes in S. cerevisiae.
Rondón AG, Mischo HE, Kawauchi J, Proudfoot NJ. Rondón AG, et al. Mol Cell. 2009 Oct 9;36(1):88-98. doi: 10.1016/j.molcel.2009.07.028. Mol Cell. 2009. PMID: 19818712 Free PMC article. - The evolutionarily conserved Pol II flap loop contributes to proper transcription termination on short yeast genes.
Pearson E, Moore C. Pearson E, et al. Cell Rep. 2014 Nov 6;9(3):821-8. doi: 10.1016/j.celrep.2014.10.007. Epub 2014 Oct 30. Cell Rep. 2014. PMID: 25437538 Free PMC article. - Quality control of mRNP in the nucleus.
Schmid M, Jensen TH. Schmid M, et al. Chromosoma. 2008 Oct;117(5):419-29. doi: 10.1007/s00412-008-0166-4. Epub 2008 Jun 18. Chromosoma. 2008. PMID: 18563427 Review. - Never a dull enzyme, RNA polymerase II.
Huang J, Ji X. Huang J, et al. Transcription. 2023 Nov;14(1-2):49-67. doi: 10.1080/21541264.2023.2208023. Epub 2023 May 2. Transcription. 2023. PMID: 37132022 Free PMC article. Review.
References
- Birse, C. E., L. Minvielle-Sebastia, B. A. Lee, W. Keller, and N. J. Proudfoot. 1998. Coupling termination of transcription to messenger RNA maturation in yeast. Science 280:298-301. - PubMed
- Buratowski, S. 2005. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 17:257-261. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases