Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae - PubMed (original) (raw)
Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae
Christophe Dez et al. EMBO J. 2006.
Erratum in
- EMBO J. 2006 Jun 7;25(11):2662
Abstract
We previously hypothesized that HEAT-repeat (Huntington, elongation A subunit, TOR) ribosome synthesis factors function in ribosome export. We report that the HEAT-repeat protein Sda1p is a component of late 60S pre-ribosomes and is required for nuclear export of both ribosomal subunits. In strains carrying the ts-lethal sda1-2 mutation, pre-60S particles were rapidly degraded following transfer to 37 degrees C. Polyadenylated forms of the 27S pre-rRNA and the 25S rRNA were detected, suggesting the involvement of the Trf4p/Air/Mtr4p polyadenylation complex (TRAMP). The absence of Trf4p suppressed polyadenylation and stabilized the pre-rRNA and rRNA. The absence of the nuclear exosome component Rrp6p also conferred RNA stabilization, with some hyperadenylation. We conclude that the nuclear-restricted pre-ribosomes are polyadenylated by TRAMP and degraded by the exosome. In sda1-2 strains at 37 degrees C, pre-40S and pre-60S ribosomes initially accumulated in the nucleoplasm, but then strongly concentrated in a subnucleolar focus, together with exosome and TRAMP components. Localization of pre-ribosomes to this focus was lost in sda1-2 strains lacking Trf4p or Rrp6p. We designate this nucleolar focus the No-body and propose that it represents a site of pre-ribosome surveillance.
Figures
Figure 1
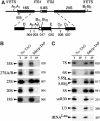
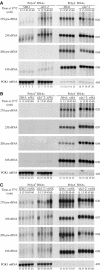
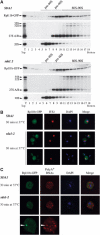
Sda1p is a component of late nuclear 60S pre-ribosomes. (A) The 35S pre-rRNA contains the sequences of the mature 18S, 5.8S and 25S rRNAs, which are separated by internal transcribed spacers 1 and 2 (ITS1 and ITS2) and flanked by the 5′ and 3′ external transcribed spacers (5′ETS and 3′ETS). Location of the different oligonucleotides used in this study is shown. (B, C) Northern analysis of rRNAs co-precipitated with Sda1-TAP or from extracts of cells lacking a tagged protein. Immunoprecipitation was performed on cell extracts using IgG-Sepharose. RNAs were extracted from the pellet after precipitation (lanes IP) or from an amount of cell extract corresponding to 1/20 (B, 1.2% denaturing agarose gel) or 1/10 (C, 8% polyacrylamide/urea gel) of the total amount used for the precipitations (lanes T). Following separation, RNAs were transferred to a nylon membrane and hybridized with the antisense oligonucleotides indicated to the right of each panel.
Figure 2
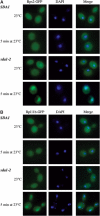
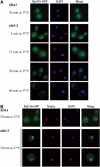
The sda1–2 mutant shows a rapid block in 40S and 60S subunit export. Analysis of 40S and 60S subunit export in sda1–2 strains expressing Rps2-GFP (A) or Rpl11b-GFP (B) reporter construct. Cells were pregrown at 23°C and then shifted to 37°C for 5 min. Cells were fixed, DAPI stained and viewed by fluorescence microscopy.
Figure 3
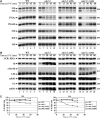
Northern analyses of pre-rRNA processing in sda1–2 mutant strains. Levels of pre-rRNAs and rRNAs were assessed by Northern blotting. Cells were pregrown at 23°C and transferred to 37°C. Samples were collected at 23°C (lanes 1, 6, 11 and 16) and after transfer to 37°C for 15 min (lanes 2, 7, 12 and 17), 30 min (lanes 3, 8, 13 and 18), 45 min (lanes 4, 9, 14 and 19) and 60 min (lanes 5, 10, 15 and 20). RNA was extracted and separated on denaturing 1.2% agarose gels (A) or on 8% polyacrylamide/urea gels (B) and transferred to a nylon membrane. Oligonucleotides used for Northern hybridization are indicated to the right of each panel (see Figure 1A for locations of oligos). (C) Quantification of data in panel A, standardized to the level of scR1 RNA.
Figure 4
Pulse-chase analyses of pre-rRNA processing in sda1–2 mutant strains. Cultures were pregrown at 23°C and transferred to 37°C for 5 min. Cells were then pulse labeled with [3H]adenine for 2 min, followed by the addition of an excess of cold adenine. Samples were collected at the times indicated, and RNAs were extracted, separated on a denaturing 1.2% agarose gel (A) or a 8% polyacrylamide/urea gel (B) and transferred to a nylon membrane. Labeled RNAs were detected by fluorography. The exposure shown for the wild-type samples is four-fold shorter than the other panels.
Figure 5
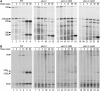
Polyadenylated rRNAs are accumulated in sda1–2 and _rrp6_Δ mutants but not in strains lacking Trf4p. The same RNA samples as used for Figure 4 were submitted to oligo(dT) selection. The resulting bound (poly(A)+ enriched) and unbound (poly(A)+ depleted) fractions were loaded on a denaturing 1.2% agarose gel and transferred to a nylon membrane. (A) Comparison of SDA1 and sda1–2 single mutants. (B) Comparison of SDA1 and sda1–2 in a _trf4_Δ background. (C) Comparison of SDA1 and sda1–2 in a _rrp6_Δ background. Oligonucleotides used for Northern hybridizations are indicated to the right of each panel. The PGK1 mRNA was used as a control for loading and polyadenylated RNA selection.
Figure 6
Rpl11p-GFP concentrates in a subnucleolar structure in sda1–2 strains 15 min after temperature shift. (A) Localization of Rpl11b-GFP in SDA1 wild-type and sda1–2 ts mutants. Aliquots were collected at different times following temperature shift and cells were fixed before they were viewed in the fluorescence microscope. (B) The Rpl11-GFP focus was visualized within the nucleolus using optical microscopy. The nucleolus was visualized using an anti-Nop1p antibody, which was subsequently recognized by an Alexafluor 555-conjugated secondary antibody.
Figure 7
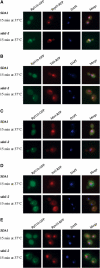
Pre-ribosome components and polyadenylated RNAs concentrate in the subnucleolar substructure. (A) Glycerol density gradient analysis of Rpl11p-GFP. A total cellular extract was prepared from wild-type and sda1.2 strains expressing Rpl11p-GFP, collected 15 min following temperature shift. Samples were loaded on a 10–30% glycerol gradient and 18 fractions were collected. Proteins and RNAs were extracted from the fractions and subjected to Western blot and Northern blot analysis, respectively. T: sample from the total unfractionated extract. (B) FISH localization of pre-rRNAs using a Cy3-labeled ITS2 probe (Leger-Silvestre et al, 2004). (C) FISH localization of polyadenylated RNAs using a Cy5-labeled oligo(dT). Wild-type and sda1.2 cells expressing Rpl11p-GFP were collected 15 min after temperature shift. Polyadenylated RNAs were imaged using a Deltavision system. These images were subjected to decovolution, and a single optical section is presented in each panel.
Figure 8
TRAMP and exosome components also concentrate in the same sharp focus in sda1–2 mutants. RFP-tagged versions of exosome (Rrp45p (A)), TRAMP (Trf4p (B), Mtr4p (C) and Air2p (D)) and proteasome (Rpn10p (E)) components were expressed in SDA1 and sda1–2 strains also expressing Rpl11-GFP. The resulting strains were pregrown at 23°C and shifted to 37°C for 15 min. Cells were fixed, DAPI stained and viewed by fluorescence microscopy.
Figure 9
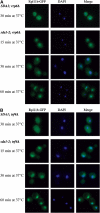
Integrity of TRAMP and exosome is required to concentrate pre-ribosomes in the subnucleolar focus. SDA1 and sda1–2 strains were deleted for RRP6 (A) or TRF4 (B) genes (see Materials and methods) and transformed with pRpl11b-GFP. The resulting strains were pregrown at 23°C and shifted to 37°C for the times indicated. Cells were fixed, DAPI stained and viewed by fluorescence microscopy.
Figure 10
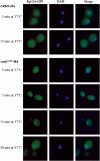
Pre-60S ribosomes form a focus in crm1-1 mutant strains. Rpl11b-GFP was expressed in strains carrying either HA-CRM1 or HA-crm1 T539C. The resulting strains were pregrown at 23°C and shifted to 37°C for the times indicated. Cells were fixed, DAPI stained and viewed by fluorescence microscopy.
Similar articles
- Has1p, a member of the DEAD-box family, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae.
Emery B, de la Cruz J, Rocak S, Deloche O, Linder P. Emery B, et al. Mol Microbiol. 2004 Apr;52(1):141-58. doi: 10.1111/j.1365-2958.2003.03973.x. Mol Microbiol. 2004. PMID: 15049817 - The splicing factor Prp43p, a DEAH box ATPase, functions in ribosome biogenesis.
Leeds NB, Small EC, Hiley SL, Hughes TR, Staley JP. Leeds NB, et al. Mol Cell Biol. 2006 Jan;26(2):513-22. doi: 10.1128/MCB.26.2.513-522.2006. Mol Cell Biol. 2006. PMID: 16382143 Free PMC article. - Immature large ribosomal subunits containing the 7S pre-rRNA can engage in translation in Saccharomyces cerevisiae.
Rodríguez-Galán O, García-Gómez JJ, Kressler D, de la Cruz J. Rodríguez-Galán O, et al. RNA Biol. 2015;12(8):838-46. doi: 10.1080/15476286.2015.1058477. RNA Biol. 2015. PMID: 26151772 Free PMC article. - Nuclear export and cytoplasmic maturation of ribosomal subunits.
Zemp I, Kutay U. Zemp I, et al. FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review. - Pre-ribosomes on the road from the nucleolus to the cytoplasm.
Tschochner H, Hurt E. Tschochner H, et al. Trends Cell Biol. 2003 May;13(5):255-63. doi: 10.1016/s0962-8924(03)00054-0. Trends Cell Biol. 2003. PMID: 12742169 Review.
Cited by
- Nucleolar localization of the yeast RNA exosome subunit Rrp44 hints at early pre-rRNA processing as its main function.
Okuda EK, Gonzales-Zubiate FA, Gadal O, Oliveira CC. Okuda EK, et al. J Biol Chem. 2020 Aug 7;295(32):11195-11213. doi: 10.1074/jbc.RA120.013589. Epub 2020 Jun 17. J Biol Chem. 2020. PMID: 32554806 Free PMC article. - The ribosome assembly factor Nop53 has a structural role in the formation of nuclear pre-60S intermediates, affecting late maturation events.
Bagatelli FFM, de Luna Vitorino FN, da Cunha JPC, Oliveira CC. Bagatelli FFM, et al. Nucleic Acids Res. 2021 Jul 9;49(12):7053-7074. doi: 10.1093/nar/gkab494. Nucleic Acids Res. 2021. PMID: 34125911 Free PMC article. - Nuclear export competence of pre-40S subunits in fission yeast requires the ribosomal protein Rps2.
Perreault A, Bellemer C, Bachand F. Perreault A, et al. Nucleic Acids Res. 2008 Nov;36(19):6132-42. doi: 10.1093/nar/gkn625. Epub 2008 Sep 27. Nucleic Acids Res. 2008. PMID: 18820293 Free PMC article. - Turnover of aberrant pre-40S pre-ribosomal particles is initiated by a novel endonucleolytic decay pathway.
Choque E, Schneider C, Gadal O, Dez C. Choque E, et al. Nucleic Acids Res. 2018 May 18;46(9):4699-4714. doi: 10.1093/nar/gky116. Nucleic Acids Res. 2018. PMID: 29481617 Free PMC article. - Regulatory mechanisms of RNA function: emerging roles of DNA repair enzymes.
Jobert L, Nilsen H. Jobert L, et al. Cell Mol Life Sci. 2014 Jul;71(13):2451-65. doi: 10.1007/s00018-014-1562-y. Epub 2014 Feb 5. Cell Mol Life Sci. 2014. PMID: 24496644 Free PMC article. Review.
References
- Andrade MA, Petosa C, O'Donoghue SI, Muller CW, Bork P (2001) Comparison of ARM and HEAT protein repeats. J Mol Biol 309: 1–18 - PubMed
- Babbio F, Farinacci M, Saracino F, Carbone ML, Privitera E (2004) Expression and localization studies of hSDA, the human ortholog of the yeast SDA1 gene. Cell Cycle 3: 486–490 - PubMed
- Baßler J, Grandi P, Gadal O, Leßmann T, Petfalski E, Tollervey D, Lechner J, Hurt E (2001) Identification of a 60S pre-ribosomal particle that is closely linked to nuclear export. Mol Cell 8: 517–529 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases