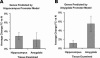Differential transcriptional response to nonassociative and associative components of classical fear conditioning in the amygdala and hippocampus - PubMed (original) (raw)
Comparative Study
. 2006 Mar-Apr;13(2):135-42.
doi: 10.1101/lm.86906. Epub 2006 Mar 17.
Affiliations
- PMID: 16547164
- PMCID: PMC1409829
- DOI: 10.1101/lm.86906
Comparative Study
Differential transcriptional response to nonassociative and associative components of classical fear conditioning in the amygdala and hippocampus
Michael B Keeley et al. Learn Mem. 2006 Mar-Apr.
Abstract
Classical fear conditioning requires the recognition of conditioned stimuli (CS) and the association of the CS with an aversive stimulus. We used Affymetrix oligonucleotide microarrays to characterize changes in gene expression compared to naive mice in both the amygdala and the hippocampus 30 min after classical fear conditioning and 30 min after exposure to the CS in the absence of an aversive stimulus. We found that in the hippocampus, levels of gene regulation induced by classical fear conditioning were not significantly greater than those induced by CS alone, whereas in the amygdala, classical fear conditioning did induce significantly greater levels of gene regulation compared to the CS. Computational studies suggest that transcriptional changes in the hippocampus and amygdala are mediated by large and overlapping but distinct combinations of molecular events. Our results demonstrate that an increase in gene regulation in the amygdala was partially correlated to associative learning and partially correlated to nonassociative components of the task, while gene regulation in the hippocampus was correlated to nonassociative components of classical fear conditioning, including configural learning.
Figures
Figure 1.
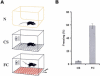
Classical fear conditioning. (A) Mice were left in their home cage (N), exposed to the conditioned stimulus (CS), or conditioned to fear the CS by coadministration of footshock (FC) and were then dissected 30 min later or tested for conditioned fear at 24 h. (B) Mice that were FC-trained demonstrated robust associative learning by exhibiting freezing behavior during ∼60% of the re-exposure to the training environment at 24 h. CS-trained mice demonstrated only baseline levels of freezing behavior.
Figure 2.
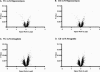
Volcano plots demonstrate the relationship between significance and Affymetrix signal ratios for gene regulation in the hippocampus and amygdala. Contrast _P_-values from a mixed model ANOVA are plotted in negative log scale on the _y_-axis against the base 2 log of the Affymetrix signal ratio for each probe set on the _x_-axis.
Figure 3.
Gene regulation was equivalent following associative learning and CS exposure in the hippocampus, whereas gene regulation in the amygdala was greater following associative learning than nonassociative learning. (A) FC versus N regulation is plotted against CS versus N regulation for each of the top 50 genes in the hippocampus (blue triangles) and amygdala (red circles). Hippocampal genes were located near the identity function, as demonstrated by the slope of the regression line (blue solid line, slope = 1.08, _R_2 = 0.96). Amygdala genes were located along a line of greater slope (red dashed line, slope = 1.54, _R_2 = 0.96). (B) Group statistics based on geometric averages across each set of 50 genes demonstrate that FC training in the hippocampus does not produce significantly greater regulation than CS training. In the amygdala, FC training does produce significantly greater regulation. Error bars represent SEM of pairwise changes.
Figure 4.
Transcription factor-binding site models predict regulation of additional genes for both amygdala and hippocampus with some specificity. (A) A set of transcription factor-binding sites identified from the 50 most significantly regulated hippocampal genes was detected in 32 additional genes, which showed significant up-regulation in the hippocampus (P < 0.001), but up-regulation in the amygdala was not significant (_P_ > 0.05). (B) A set of transcription factor-binding sites identified from the 50 most significantly regulated amygdala genes was detected in 27 additional genes, which showed significant up-regulation in the amygdala (P < 0.005), whereas up-regulation in the hippocampus was not significant (_P_ > 0.05).
Similar articles
- Elevated Arc/Arg 3.1 protein expression in the basolateral amygdala following auditory trace-cued fear conditioning.
Chau LS, Prakapenka A, Fleming SA, Davis AS, Galvez R. Chau LS, et al. Neurobiol Learn Mem. 2013 Nov;106:127-33. doi: 10.1016/j.nlm.2013.07.010. Epub 2013 Jul 24. Neurobiol Learn Mem. 2013. PMID: 23891993 - Differential contribution of amygdala and hippocampus to cued and contextual fear conditioning.
Phillips RG, LeDoux JE. Phillips RG, et al. Behav Neurosci. 1992 Apr;106(2):274-85. doi: 10.1037//0735-7044.106.2.274. Behav Neurosci. 1992. PMID: 1590953 - Consolidation of CS and US representations in associative fear conditioning.
Frankland PW, Josselyn SA, Anagnostaras SG, Kogan JH, Takahashi E, Silva AJ. Frankland PW, et al. Hippocampus. 2004;14(5):557-69. doi: 10.1002/hipo.10208. Hippocampus. 2004. PMID: 15301434 - Neurobiology of Pavlovian fear conditioning.
Maren S. Maren S. Annu Rev Neurosci. 2001;24:897-931. doi: 10.1146/annurev.neuro.24.1.897. Annu Rev Neurosci. 2001. PMID: 11520922 Review. - What the amygdala does and doesn't do in aversive learning.
Maren S. Maren S. Learn Mem. 2003 Sep-Oct;10(5):306-8. doi: 10.1101/lm.68403. Learn Mem. 2003. PMID: 14557601 Review. No abstract available.
Cited by
- Molecular mechanisms of fear learning and memory.
Johansen JP, Cain CK, Ostroff LE, LeDoux JE. Johansen JP, et al. Cell. 2011 Oct 28;147(3):509-24. doi: 10.1016/j.cell.2011.10.009. Cell. 2011. PMID: 22036561 Free PMC article. Review. - The Molecular Substrates of Second-Order Conditioned Fear in the Basolateral Amygdala Complex.
Leake J, Shvetcov A, Clemens KJ, Kershaw K, Westbrook RF, Holmes NM. Leake J, et al. J Neurosci. 2025 Jun 18;45(25):e1087242025. doi: 10.1523/JNEUROSCI.1087-24.2025. J Neurosci. 2025. PMID: 40345834 - Memory acquisition and retrieval impact different epigenetic processes that regulate gene expression.
Peixoto LL, Wimmer ME, Poplawski SG, Tudor JC, Kenworthy CA, Liu S, Mizuno K, Garcia BA, Zhang NR, Giese K, Abel T. Peixoto LL, et al. BMC Genomics. 2015;16 Suppl 5(Suppl 5):S5. doi: 10.1186/1471-2164-16-S5-S5. Epub 2015 May 26. BMC Genomics. 2015. PMID: 26040834 Free PMC article. - Long-term memory deficits in Pavlovian fear conditioning in Ca2+/calmodulin kinase kinase alpha-deficient mice.
Blaeser F, Sanders MJ, Truong N, Ko S, Wu LJ, Wozniak DF, Fanselow MS, Zhuo M, Chatila TA. Blaeser F, et al. Mol Cell Biol. 2006 Dec;26(23):9105-15. doi: 10.1128/MCB.01452-06. Epub 2006 Oct 2. Mol Cell Biol. 2006. PMID: 17015467 Free PMC article. - The microbiome regulates amygdala-dependent fear recall.
Hoban AE, Stilling RM, Moloney G, Shanahan F, Dinan TG, Clarke G, Cryan JF. Hoban AE, et al. Mol Psychiatry. 2018 May;23(5):1134-1144. doi: 10.1038/mp.2017.100. Epub 2017 May 16. Mol Psychiatry. 2018. PMID: 28507320 Free PMC article.
References
- Abel T., Lattal K.M. Molecular mechanisms of memory acquisition, consolidation and retrieval. Curr. Opin. Neurobiol. 2001;11:180–187. - PubMed
- Barrientos R.M., O’Reilly R.C., Rudy J.W. Memory for context is impaired by injecting anisomycin into dorsal hippocampus following context exploration. Behav. Brain Res. 2002;134:299–306. - PubMed
- Bernabeu R., Bevilaqua L., Ardenghi P., Bromberg E., Schmitz P., Bianchin M., Izquierdo I., Medina J.H. Involvement of hippocampal cAMP/cAMP-dependent protein kinase signaling pathways in a late memory consolidation phase of aversively motivated learning in rats. Proc. Natl. Acad. Sci. 1997;94:7041–7046. - PMC - PubMed
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases