An application of statistics to comparative metagenomics - PubMed (original) (raw)
Comparative Study
An application of statistics to comparative metagenomics
Beltran Rodriguez-Brito et al. BMC Bioinformatics. 2006.
Abstract
Background: Metagenomics, sequence analyses of genomic DNA isolated directly from the environments, can be used to identify organisms and model community dynamics of a particular ecosystem. Metagenomics also has the potential to identify significantly different metabolic potential in different environments.
Results: Here we use a statistical method to compare curated subsystems, to predict the physiology, metabolism, and ecology from metagenomes. This approach can be used to identify those subsystems that are significantly different between metagenome sequences. Subsystems that were overrepresented in the Sargasso Sea and Acid Mine Drainage metagenome when compared to non-redundant databases were identified.
Conclusion: The methodology described herein applies statistics to the comparisons of metabolic potential in metagenomes. This analysis reveals those subsystems that are more, or less, represented in the different environments that are compared. These differences in metabolic potential lead to several testable hypotheses about physiology and metabolism of microbes from these ecosystems.
Figures
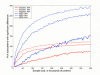
Figure 1
Effect of sample size on identifying differences between phylosubsystems. The red lines reflect the number of phylosubsystems overrepresented in the Sargasso Sea dataset. The blue lines represented the number of phylosubsystems overrepresented in the SEED dataset. Three different confidence levels (90, 95, and 99%) are plotted.
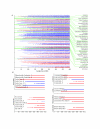
Figure 2
Significantly different subsystems between Sargasso Sea and SEED datasets. (A). Each subsystem was bootstrapped with between 10 and 400 samples per bootstrap, and subsystems that are significantly different with 99% and 2,000 bootstraps are highlighted. Those subsystems that are significantly more prevalent in the SEED database are colored blue, and those subsystems that are significantly more prevalent in the Sargasso Sea dataset are colored red. (B). Magnified view of several different subsystems. Subsystems from amino acid synthesis, carbohydrate utilization, cofactor synthesis, fatty acids, nucleotide synthesis, and photosynthesis are shown in more detail. Colors are as described for (A). E = eukaryotic subsystem, B = bacterial subsystem, A = archaeal subsystem.
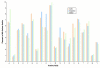
Figure 3
Fraction of amino acids in metagenomes. The fraction of each amino acid in all the predicted proteins in the three data samples was counted and compared.
Figure 4
Flow chart of methods used to identify statistical differences between phylosubsystems.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources