Molecular functions of nuclear actin in transcription - PubMed (original) (raw)
Review
Molecular functions of nuclear actin in transcription
Piergiorgio Percipalle et al. J Cell Biol. 2006.
Abstract
Actin is not only a major cytoskeletal component in all eukaryotic cells but also a nuclear protein that plays a role in gene transcription. We put together data from in vitro and in vivo experiments that begin to provide insights into the molecular mechanisms by which actin functions in transcription. Recent studies performed in vitro have suggested that actin, in direct contact with the transcription apparatus, is required in an early step of transcription that is common to all three eukaryotic RNA polymerases. In addition, there is evidence from in vivo studies that actin is involved in the transcription elongation of class II genes. In this case, actin is bound to a specific subset of premessenger RNA binding proteins, and the actin-messenger RNP complex may constitute a molecular platform for recruitment of histone-modifying enzymes. We discuss a general model for actin in RNA polymerase II transcription whereby actin works as a conformational switch in conjunction with specific adaptors to facilitate the remodeling of large macromolecular assemblies at the promoter and along the active gene.
Figures
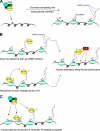
Figure 1.
Models for the function of actin in chromatin regulation at class II genes. (A) Actin is a component of ATP-dependent chromatin remodeling complexes (CRCs) involved in transcriptional activation. (B) Actin becomes incorporated into the growing pre-mRNP while transcription is taking place. Actin may be recruited directly by hnRNP proteins. An alternative mechanism, supported by the observed binding of actin to the CTD, is that actin is recruited to the transcription machinery via binding to the CTD and subsequently transferred from pol-II to the nascent transcript. Adaptor proteins, such as HRP65-2 in C. tentans and hnRNP U in mammals (see the text for details), help actin recruit HATs that acetylate histones along the gene and maintain the chromatin template in a transcription-competent state. (C) We speculate that actin mediates the cotranscriptional recruitment of chromatin remodeling complexes to active genes through interactions with pol-II and/or with the nascent pre-mRNP.
Similar articles
- Actin in transcription and transcription regulation.
Miralles F, Visa N. Miralles F, et al. Curr Opin Cell Biol. 2006 Jun;18(3):261-6. doi: 10.1016/j.ceb.2006.04.009. Epub 2006 May 9. Curr Opin Cell Biol. 2006. PMID: 16687246 Review. - Actin and myosin as transcription factors.
Grummt I. Grummt I. Curr Opin Genet Dev. 2006 Apr;16(2):191-6. doi: 10.1016/j.gde.2006.02.001. Epub 2006 Feb 21. Curr Opin Genet Dev. 2006. PMID: 16495046 Review. - Transcriptional control of gene expression by actin and myosin.
Louvet E, Percipalle P. Louvet E, et al. Int Rev Cell Mol Biol. 2009;272:107-47. doi: 10.1016/S1937-6448(08)01603-1. Int Rev Cell Mol Biol. 2009. PMID: 19121817 Review. - A mechanism for coordinating chromatin modification and preinitiation complex assembly.
Black JC, Choi JE, Lombardo SR, Carey M. Black JC, et al. Mol Cell. 2006 Sep 15;23(6):809-18. doi: 10.1016/j.molcel.2006.07.018. Mol Cell. 2006. PMID: 16973433 - Chromatin-associated regulation of HIV-1 transcription: implications for the development of therapeutic strategies.
Quivy V, De Walque S, Van Lint C. Quivy V, et al. Subcell Biochem. 2007;41:371-96. Subcell Biochem. 2007. PMID: 17484137 Review.
Cited by
- Nuclear Actin Dynamics in Gene Expression, DNA Repair, and Cancer.
Huang Y, Zhang S, Park JI. Huang Y, et al. Results Probl Cell Differ. 2022;70:625-663. doi: 10.1007/978-3-031-06573-6_23. Results Probl Cell Differ. 2022. PMID: 36348125 Free PMC article. - Moving chromatin within the interphase nucleus-controlled transitions?
Chuang CH, Belmont AS. Chuang CH, et al. Semin Cell Dev Biol. 2007 Oct;18(5):698-706. doi: 10.1016/j.semcdb.2007.08.012. Epub 2007 Aug 25. Semin Cell Dev Biol. 2007. PMID: 17905613 Free PMC article. Review. - Identification of FBXO25-interacting proteins using an integrated proteomics approach.
Teixeira FR, Yokoo S, Gartner CA, Manfiolli AO, Baqui MM, Assmann EM, Maragno AL, Yu H, de Lanerolle P, Kobarg J, Gygi SP, Gomes MD. Teixeira FR, et al. Proteomics. 2010 Aug;10(15):2746-57. doi: 10.1002/pmic.200900419. Proteomics. 2010. PMID: 20473970 Free PMC article. - Actin: its cumbersome pilgrimage through cellular compartments.
Schleicher M, Jockusch BM. Schleicher M, et al. Histochem Cell Biol. 2008 Jun;129(6):695-704. doi: 10.1007/s00418-008-0430-y. Epub 2008 Apr 26. Histochem Cell Biol. 2008. PMID: 18438682 Free PMC article. Review.
References
- Amankwah, K.S., and U. de Boni. 1994. Ultrastructural localization of filamentous actin within neuronal interphase nuclei in situ. Exp. Cell Res. 210:315–325. - PubMed
- Andersen, J.S., C.E. Lyon, A.H. Fox, A.K. Leung, Y.W. Lam, H. Steen, M. Mann, and A.I. Lamond. 2002. Directed proteomic analysis of the human nucleolus. Curr. Biol. 12:1–11. - PubMed
- Andersen, J.S., Y.W. Lam, A.K. Leung, S.E. Ong, C.E. Lyon, A.I. Lamond, and M. Mann. 2005. Nucleolar proteome dynamics. Nature. 433:77–83. - PubMed
- Bettinger, B.T., D.M. Gilbert, and D.C. Amberg. 2004. Actin up in the nucleus. Nat. Rev. Mol. Cell Biol. 5:410–415. - PubMed
- Boyer, L.A., and C.L. Peterson. 2000. Actin-related proteins (Arps): conformational switches for chromatin-remodeling machines? Bioessays. 22:666–672. - PubMed