Experimental determination of the radiation dose limit for cryocooled protein crystals - PubMed (original) (raw)
Experimental determination of the radiation dose limit for cryocooled protein crystals
Robin Leslie Owen et al. Proc Natl Acad Sci U S A. 2006.
Abstract
Radiation damage to cryocooled protein crystals during x-ray structure determination has become an inherent part of macromolecular diffraction data collection at third-generation synchrotrons. Generally, radiation damage is an undesirable component of the experiment and can result in erroneous structural detail in the final model. The characterization of radiation damage thus has become an important area for structural biologists. The calculated dose limit of 2 x 10(7) Gy for the diffracting power of cryocooled protein crystals to drop by half has been experimentally evaluated at a third-generation synchrotron source. Successive data sets were collected from four holoferritin and three apoferritin crystals. The absorbed dose for each crystal was calculated by using the program raddose after measurement of the incident photon flux and determination of the elemental crystal composition by micro-particle-induced x-ray emission. Degradation in diffraction quality and specific structural changes induced by synchrotron radiation then could be compared directly with absorbed dose for different dose/dose rate regimes: a 10% lifetime decrease for a 10-fold dose rate increase was observed. Remarkable agreement both between different crystals of the same type and between apoferritin and holoferritin was observed for the dose required to reduce the diffracted intensity by half (D(1/2)). From these measurements, a dose limit of D(1/2) = 4.3 (+/-0.3) x10(7) Gy was obtained. However, by considering other data quality indicators, an intensity reduction to I(ln2) = ln2 x I(0), corresponding to an absorbed dose of 3.0 x 10(7) Gy, is recommended as an appropriate dose limit for typical macromolecular crystallography experiments.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
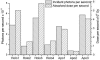
Fig. 1.
Bar chart showing the incident photons per second and the absorbed dose per second for the four holoferritin and three apoferritin crystals studied. The difference in the relative size of the photons per second bar compared with the dose per second bar for the two crystal types highlights the necessity of taking careful account of the crystal composition when calculating the absorbed dose.
Fig. 2.
Elemental areal concentration microPIXE maps (500 × 500 μm) obtained by scanning a 1-μm-diameter proton beam in x and y across a holoferritin crystal (holo1): sulfur (Left), iron (Center), and cadmium (Right) distributions. The iron is localized in the protein crystal, whereas because of the presence of ammonium sulfate in the solvent, the sulfur is more spread out.
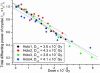
Fig. 3.
Graph showing the total diffracting power of four different holoferritin crystals as a function of absorbed dose. The sum of the intensities of all reflections, j, Σ_I_ j have been normalized to the intensity of the first data set [_I_0] for each crystal, giving ΣTot = _I_D/_I_0. The results show clear agreement between the dose required to reduce the diffracting intensity by half (_D_1/2) for each of the crystals.
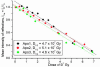
Fig. 4.
Graph showing the total normalized diffracting power, _Ī_Tot, of three different apoferritin crystals as a function of absorbed dose. The results show both agreement between the dose required to reduce the diffracting intensity by half (_D_1/2) for the different crystals and also for the values of _D_1/2 obtained for holoferritin.
Fig. 5.
Electron density of the structures (contoured at 0.2 electrons per Å3) refined from the first eight data sets (Top and Middle) of the holo3 crystal, showing the specific structural damage suffered by Glu-63, Arg-52 (from a symmetry-related molecule), and a water molecule. Bottom shows residue Thr-29 from every other data set. It shows no degradation in the quality of the electron density, similar to the sulfur-containing residue (Cys-48) visible in the top eight maps. The resilience of these residues to high dose illustrates the highly specific nature of radiation damage in proteins.
Fig. 6.
Bar chart showing the number of damaged residues for each crystal vs. absorbed dose. The dotted vertical line indicates the experimentally determined value of _D_1/2. Note that there are no disulfide bridges in ferritin, and these are known to be the most susceptible bonds.
Similar articles
- Reduction of X-ray-induced radiation damage of macromolecular crystals by data collection at 15 K: a systematic study.
Meents A, Wagner A, Schneider R, Pradervand C, Pohl E, Schulze-Briese C. Meents A, et al. Acta Crystallogr D Biol Crystallogr. 2007 Mar;63(Pt 3):302-9. doi: 10.1107/S0907444906053261. Epub 2007 Feb 21. Acta Crystallogr D Biol Crystallogr. 2007. PMID: 17327667 - Radiation decay of thaumatin crystals at three X-ray energies.
Liebschner D, Rosenbaum G, Dauter M, Dauter Z. Liebschner D, et al. Acta Crystallogr D Biol Crystallogr. 2015 Apr;71(Pt 4):772-8. doi: 10.1107/S1399004715001030. Epub 2015 Mar 26. Acta Crystallogr D Biol Crystallogr. 2015. PMID: 25849388 Free PMC article. - Know your dose: RADDOSE.
Paithankar KS, Garman EF. Paithankar KS, et al. Acta Crystallogr D Biol Crystallogr. 2010 Apr;66(Pt 4):381-8. doi: 10.1107/S0907444910006724. Epub 2010 Mar 24. Acta Crystallogr D Biol Crystallogr. 2010. PMID: 20382991 Free PMC article. - Radiation Damage in Macromolecular Crystallography.
Garman EF, Weik M. Garman EF, et al. Methods Mol Biol. 2017;1607:467-489. doi: 10.1007/978-1-4939-7000-1_20. Methods Mol Biol. 2017. PMID: 28573586 Review.
Cited by
- Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Huang CY, Aumonier S, Olieric V, Wang M. Huang CY, et al. Acta Crystallogr D Struct Biol. 2024 Aug 1;80(Pt 8):620-628. doi: 10.1107/S2059798324006697. Epub 2024 Jul 25. Acta Crystallogr D Struct Biol. 2024. PMID: 39052318 Free PMC article. - Doses for X-ray and electron diffraction: New features in RADDOSE-3D including intensity decay models.
Dickerson JL, McCubbin PTN, Brooks-Bartlett JC, Garman EF. Dickerson JL, et al. Protein Sci. 2024 Jul;33(7):e5005. doi: 10.1002/pro.5005. Protein Sci. 2024. PMID: 38923423 Free PMC article. - Crystal structure via fluctuation scattering.
Adams P, Greaves TL, Martin AV. Adams P, et al. IUCrJ. 2024 Jul 1;11(Pt 4):538-555. doi: 10.1107/S2052252524003932. IUCrJ. 2024. PMID: 38842120 Free PMC article. - Identifying and avoiding radiation damage in macromolecular crystallography.
Shelley KL, Garman EF. Shelley KL, et al. Acta Crystallogr D Struct Biol. 2024 May 1;80(Pt 5):314-327. doi: 10.1107/S2059798324003243. Epub 2024 Apr 30. Acta Crystallogr D Struct Biol. 2024. PMID: 38700059 Free PMC article. - Characterization of Biological Samples Using Ultra-Short and Ultra-Bright XFEL Pulses.
Round A, Jungcheng E, Fortmann-Grote C, Giewekemeyer K, Graceffa R, Kim C, Kirkwood H, Mills G, Round E, Sato T, Pascarelli S, Mancuso A. Round A, et al. Adv Exp Med Biol. 2024;3234:141-162. doi: 10.1007/978-3-031-52193-5_10. Adv Exp Med Biol. 2024. PMID: 38507205
References
- Blake C., Phillips D. C. Proceedings of the Symposium on the Biological Effects of Ionizing Radiation at the Molecular Level; Vienna: International Atomic Energy Agency; 1962. pp. 183–191.
- Haas D., Rossmann M. G. Acta Crystallogr. B. 1970;26:998–1004. - PubMed
- Garman E. F., Schneider T. R. J. Appl. Cryst. 1997;27:211–237.
- Rodgers D. W. Methods Enzymol. 1997;276:183–202. - PubMed
- Burmeister W. P. Acta Crystallogr. D. 2000;56:328–341. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources