Systematic identification of microRNA functions by combining target prediction and expression profiling - PubMed (original) (raw)
Systematic identification of microRNA functions by combining target prediction and expression profiling
Xiaowei Wang et al. Nucleic Acids Res. 2006.
Abstract
Target predictions and validations are major obstacles facing microRNA (miRNA) researchers. Animal miRNA target prediction is challenging because of limited miRNA sequence complementarity to the targets. In addition, only a small number of predicted targets have been experimentally validated and the miRNA mechanism is poorly understood. Here we present a novel algorithm for animal miRNA target prediction. The algorithm combines relevant parameters for miRNA target recognition and heuristically assigns different weights to these parameters according to their relative importance. A score calculation scheme is introduced to reflect the strength of each parameter. We also performed microarray time course experiments to identify downregulated genes due to miRNA overexpression. The computational target prediction is combined with the miRNA transfection experiment to systematically identify the gene targets of human miR-124. miR-124 overexpression led to a significant downregulation of many cell cycle related genes. This may be the result of direct suppression of a few cell growth inhibitors at the early stage of miRNA overexpression, and these targeted genes were continuously suppressed over a long period of time. Our high-throughput approach can be generalized to globally identify the targets and functions of other miRNAs.
Figures
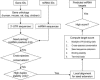
Figure 1
A simple flowchart for miRNA target prediction algorithm.
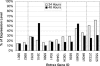
Figure 2
Real-time RT–PCR validation of the downregulated predicted miR-124 targets. The mRNA levels for downregulated targets at 4, 24 and 48 h were examined. The expression levels at 24 and 48 h are presented as percentages of that at 4 h. Among the validated targets, the microarray results indicated that genes 8801 and 8992 were first downregulated at 8 h; genes 8763, 3915, 6566, 9341, 10 449 and 10 634 were first downregulated at 16 h; genes 6836, 60 481 and 84 061 were first downregulated at 24 h; genes 27 230 and 55 225 were first downregulated at 32 h (downregulation is defined as at least 50% reduction of gene expression).
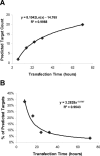
Figure 3
miR-124 overexpression time course analysis to identify predicted targets in total downregulated genes. (A) Counts of downregulated predicted targets at different time points after miR-124 transfections. (B) The percentages of predicted targets in total downregulated genes.
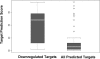
Figure 4
A boxplot to compare the prediction scores from all predicted miR-124 targets and from downregulated targets 24 h after transfection. The box in each panel represents the 50% range of each target list. The median scores were 33 and 54 for all human predicted targets and the downregulated targets, respectively.
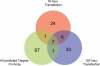
Figure 5
Overlapping downregulated genes after 16 and 120 h of miR-124 overexpression. Thirteen genes were in both lists and seven of them were miR-124 targets. Overall, 76 predicted targets were represented on the arrays.
Similar articles
- MicroRNA target prediction by expression analysis of host genes.
Gennarino VA, Sardiello M, Avellino R, Meola N, Maselli V, Anand S, Cutillo L, Ballabio A, Banfi S. Gennarino VA, et al. Genome Res. 2009 Mar;19(3):481-90. doi: 10.1101/gr.084129.108. Epub 2008 Dec 16. Genome Res. 2009. PMID: 19088304 Free PMC article. - Characterization of human platelet microRNA by quantitative PCR coupled with an annotation network for predicted target genes.
Osman A, Fälker K. Osman A, et al. Platelets. 2011;22(6):433-41. doi: 10.3109/09537104.2011.560305. Epub 2011 Mar 25. Platelets. 2011. PMID: 21438667 - Human microRNA target identification by RRSM.
Hsieh WJ, Wang H. Hsieh WJ, et al. J Theor Biol. 2011 Oct 7;286(1):79-84. doi: 10.1016/j.jtbi.2011.06.022. Epub 2011 Jun 29. J Theor Biol. 2011. PMID: 21736879 - Bayesian inference of MicroRNA targets from sequence and expression data.
Huang JC, Morris QD, Frey BJ. Huang JC, et al. J Comput Biol. 2007 Jun;14(5):550-63. doi: 10.1089/cmb.2007.R002. J Comput Biol. 2007. PMID: 17683260 Review. - Desperately seeking microRNA targets.
Thomas M, Lieberman J, Lal A. Thomas M, et al. Nat Struct Mol Biol. 2010 Oct;17(10):1169-74. doi: 10.1038/nsmb.1921. Nat Struct Mol Biol. 2010. PMID: 20924405 Review.
Cited by
- Identification of candidate regulatory sequences in mammalian 3' UTRs by statistical analysis of oligonucleotide distributions.
Corà D, Di Cunto F, Caselle M, Provero P. Corà D, et al. BMC Bioinformatics. 2007 May 24;8:174. doi: 10.1186/1471-2105-8-174. BMC Bioinformatics. 2007. PMID: 17524134 Free PMC article. - miRDB: a microRNA target prediction and functional annotation database with a wiki interface.
Wang X. Wang X. RNA. 2008 Jun;14(6):1012-7. doi: 10.1261/rna.965408. Epub 2008 Apr 21. RNA. 2008. PMID: 18426918 Free PMC article. - Identification and target prediction of miRNAs specifically expressed in rat neural tissue.
Hua YJ, Tang ZY, Tu K, Zhu L, Li YX, Xie L, Xiao HS. Hua YJ, et al. BMC Genomics. 2009 May 9;10:214. doi: 10.1186/1471-2164-10-214. BMC Genomics. 2009. PMID: 19426523 Free PMC article. - Global identification of microRNAs associated with chlorantraniliprole resistance in diamondback moth Plutella xylostella (L.).
Zhu B, Li X, Liu Y, Gao X, Liang P. Zhu B, et al. Sci Rep. 2017 Jan 18;7:40713. doi: 10.1038/srep40713. Sci Rep. 2017. PMID: 28098189 Free PMC article. - Improving microRNA target prediction by modeling with unambiguously identified microRNA-target pairs from CLIP-ligation studies.
Wang X. Wang X. Bioinformatics. 2016 May 1;32(9):1316-22. doi: 10.1093/bioinformatics/btw002. Epub 2016 Jan 6. Bioinformatics. 2016. PMID: 26743510 Free PMC article.
References
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
- He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nature Rev. Genet. 2004;5:522–531. - PubMed
- Miska E.A. How microRNAs control cell division, differentiation and death. Curr. Opin. Genet. Dev. 2005;15:563–568. - PubMed
- O'Donnell K.A., Wentzel E.A., Zeller K.I., Dang C.V., Mendell J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. - PubMed
- Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K.L., Brown D., Slack F.J. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases