Rho-dependent transcription termination: more questions than answers - PubMed (original) (raw)
Review
Rho-dependent transcription termination: more questions than answers
Sharmistha Banerjee et al. J Microbiol. 2006 Feb.
Abstract
Escherichia coli protein Rho is required for the factor-dependent transcription termination by an RNA polymerase and is essential for the viability of the cell. It is a homohexameric protein that recognizes and binds preferably to C-rich sites in the transcribed RNA. Once bound to RNA, it utilizes RNA-dependent ATPase activity and subsequently ATPase-dependent helicase activity to unwind RNA-DNA hybrids and release RNA from a transcribing elongation complex. Studies over the past few decades have highlighted Rho as a molecule and have revealed much of its mechanistic properties. The recently solved crystal structure could explain many of its physiological functions in terms of its structure. Despite all these efforts, many of the fundamental questions pertaining to Rho recognition sites, differential ATPase activity in response to different RNAs, translocation of Rho along the nascent transcript, interactions with elongation complex and finally unwinding and release of RNA remain obscure. In the present review we have attempted to summarize "the knowns" and "the unknowns" of the Rho protein revealed by the recent developments in this field. An attempt has also been made to understand the physiology of Rho in the light of its phylogeny.
Figures
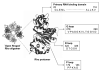
Fig. 1
Functional domains of a Rho protomer of E. coli Rho. The figure shows an open ‘key washer’ like pentameric form of Rho. Each protomer has four defined functional domains: Primary RNA binding domain that can bind to a single stranded DNA molecule as well as single stranded RNA molecule extending from residues 22 to 116; P-loop, for ATP binding and ATPase activity, includes residues from 179-183; Q-loop, from 278-290 and R-loop from 322-326 comprising the secondary RNA binding site of Rho.
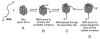
Fig. 2
An overview of the steps involved in the termination by Rho. The exact phenomenon of Rho translocation, interaction with elongation complex and finally unwinding and releasing RNA are still ambiguous.
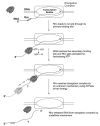
Fig. 3
Events of Rho loading and formation of ATPase competent intermediates (based on Kim and Patel 2001). RNA randomly collides with the Rho molecule forming a dynamic Rho-RNA complex. RNA then wraps around the primary binding site of each of the five protomer of the Rho open complex forming a stable moiety. The RNA subsequently passes towards the center of the open Rho ring making contact with the secondary binding site of the Rho molecule forming a steady complex. Once RNA enters the ‘ring’ of Rho, the open conformation gets closed by a sixth subunit, leading to the formation of a highly stable ATPase competent moiety.
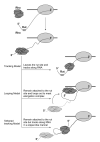
Fig. 4
Translocation models of Rho along the template. Tracking, looping, and tethered tracking models.
Fig. 5
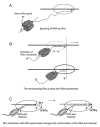
Models to explain Rho’s access to RNA-DNA hybrid and release RNA. A, it spools RNA from a distance without making contact with elongation complex. B, Rho forward translocates the elongation complex to get access to the RNA. C, Rho contact with elongation complex changes the conformation of RNA exit channel.
Fig. 6
Schematic representation of the conserved domains of E. coli Rho through NCBI Blast. a. Rho-factor: Transcription termination factor Rho is a bacterial ATP-dependent RNA/DNA helicase. b. ATPase-fllagellum or Flagellum-specific ATPase/type III secretory pathway virulence-related protein: This group of ATPases are responsible for the export of flagellum and virulence-related proteins. The bacterial flagellar motor is similar to the F0F1-ATPase, in that they both are proton driven rotary molecular devices. c. RecA-like NTPases: This family includes the NTP binding domain of F1 and V1 H+ATPases, DnaB and related helicases as well as bacterial RecA and related eukaryotic and archaeal recombinases. d. F1 ATP synthase beta subunit, nucleotide-binding domain: The F-ATPase is found in bacterial plasma membranes, mitochondrial inner membranes and in chloroplast thylakoid membranes. It uses a proton gradient to drive ATP synthesis and hydrolyzes ATP to build the proton gradient. e. V/A-type ATP synthase catalytic subunit A. These ATPases couple ATP hydrolysis to the build up of a H+ gradient, but V-type ATPases do not catalyze the reverse reaction. The Vacuolar (V-type) ATPase is found in the membranes of vacuoles, the golgi apparatus and in other coated vesicles in eukaryotes. f. F1 ATP synthase alpha, central domain. The F-ATPase is found in bacterial plasma membranes, mitochondrial inner membranes and in chloroplast thylakoid membranes g. CSP, Cold shock protein domain; RNA-binding domain that functions as a RNA-chaperone in bacteria and is involved in regulating translation in eukaryotes. Contains sub-family of RNA-binding domains in the Rho transcription termination factor. h. AAA-ATPases associated with a variety of cellular activities. i. ATP synthase alpha/beta family, nucleotide-binding domain. This family includes the ATP synthase alpha and beta subunits the ATP synthase associated with flagella. j. Rho: Transcription termination factor. k. Flagellar biosynthesis/type III secretory pathway ATPase: involved in cell motility and secretion/Intracellular trafficking and secretion. l. AtpA: F0F1-type ATP synthase, alpha subunit involved in energy production and conversion. m. NtpA: Archaeal/vacuolar-type H+-ATPase subunit A involved in energy production and conversion.
Similar articles
- The mechanism of early transcription termination by Rho026.
Washburn RS, Jin DJ, Stitt BL. Washburn RS, et al. J Mol Biol. 1996 Jul 19;260(3):347-58. doi: 10.1006/jmbi.1996.0405. J Mol Biol. 1996. PMID: 8757798 - Interaction with the nascent RNA is a prerequisite for the recruitment of Rho to the transcription elongation complex in vitro.
Kalyani BS, Muteeb G, Qayyum MZ, Sen R. Kalyani BS, et al. J Mol Biol. 2011 Oct 28;413(3):548-60. doi: 10.1016/j.jmb.2011.08.053. Epub 2011 Sep 6. J Mol Biol. 2011. PMID: 21920369 - Kinetics of the RNA-DNA helicase activity of Escherichia coli transcription termination factor rho. 2. Processivity, ATP consumption, and RNA binding.
Walstrom KM, Dozono JM, von Hippel PH. Walstrom KM, et al. Biochemistry. 1997 Jul 1;36(26):7993-8004. doi: 10.1021/bi963180r. Biochemistry. 1997. PMID: 9201946 - Rho and RNA: models for recognition and response.
Platt T. Platt T. Mol Microbiol. 1994 Mar;11(6):983-90. doi: 10.1111/j.1365-2958.1994.tb00376.x. Mol Microbiol. 1994. PMID: 8022288 Review. - Keeping up to speed with the transcription termination factor Rho motor.
Boudvillain M, Nollmann M, Margeat E. Boudvillain M, et al. Transcription. 2010 Sep-Oct;1(2):70-5. doi: 10.4161/trns.1.2.12232. Transcription. 2010. PMID: 21326894 Free PMC article. Review.
Cited by
- Rho and NusG suppress pervasive antisense transcription in Escherichia coli.
Peters JM, Mooney RA, Grass JA, Jessen ED, Tran F, Landick R. Peters JM, et al. Genes Dev. 2012 Dec 1;26(23):2621-33. doi: 10.1101/gad.196741.112. Genes Dev. 2012. PMID: 23207917 Free PMC article. - YihE is a novel binding partner of Rho and regulates Rho-dependent transcription termination in the Cpx stress response.
Wang B, Pei H, Lu Z, Xu Y, Han S, Jia Z, Zheng J. Wang B, et al. iScience. 2022 Nov 2;25(12):105483. doi: 10.1016/j.isci.2022.105483. eCollection 2022 Dec 22. iScience. 2022. PMID: 36387024 Free PMC article. - ADP but not P(i) dissociation contributes to rate limitation for Escherichia coli Rho.
Chen X, Stitt BL. Chen X, et al. J Biol Chem. 2009 Dec 4;284(49):33773-80. doi: 10.1074/jbc.M109.056473. Epub 2009 Oct 16. J Biol Chem. 2009. PMID: 19837672 Free PMC article. - RNA secondary structures regulate three steps of Rho-dependent transcription termination within a bacterial mRNA leader.
Kriner MA, Groisman EA. Kriner MA, et al. Nucleic Acids Res. 2017 Jan 25;45(2):631-642. doi: 10.1093/nar/gkw889. Epub 2016 Oct 5. Nucleic Acids Res. 2017. PMID: 28123036 Free PMC article. - The Rho-Dependent Transcription Termination Is Involved in Broad-Spectrum Antibiotic Susceptibility in Escherichia coli.
Hafeezunnisa M, Sen R. Hafeezunnisa M, et al. Front Microbiol. 2020 Nov 30;11:605305. doi: 10.3389/fmicb.2020.605305. eCollection 2020. Front Microbiol. 2020. PMID: 33329496 Free PMC article.
References
- Adhya S, Gottesman M. Control of transcription termination. Annu. Rev. Biochem. 1978;47:967–96. - PubMed
- Alifano P, Rivellini F, Limauro D, Bruni CB, Carlomagno MS. A consensus motif common to all Rho-dependent prokaryotic transcription terminators. Cell. 1991;64:553–563. - PubMed
- Bear DG, Hicks PS, Escudero KW, Andrews CL, McSwiggen JA, von Hippel PH. Interactions of Escherichia coli transcription termination factor Rho with RNA. II. Electron microscopy and nuclease protection experiments. J. Mol. Biol. 1988;199:623–635. - PubMed
- Bogden CE, Fass D, Bergman N, Nichols MD, Berger JM. The structural basis for terminator recognition by the Rho transcription termination factor. Mol. Cell. 1999;3:487–493. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous