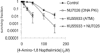Inhibition of poly (ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair - PubMed (original) (raw)
Inhibition of poly (ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair
Helen E Bryant et al. Nucleic Acids Res. 2006.
Abstract
Poly (ADP-ribose) polymerase (PARP-1), ATM and DNA-dependent protein kinase (DNA-PK) are all involved in responding to DNA damage to activate pathways responsible for cellular survival. Here, we demonstrate that PARP-1-/- cells are sensitive to the ATM inhibitor KU55933 and conversely that AT cells are sensitive to the PARP inhibitor 4-amino-1,8-napthalamide. In addition, PARP-1-/- cells are shown to be sensitive to the DNA-PK inhibitor NU7026 and DNA-PKcs or Ku80 defective cells shown to be sensitive to PARP inhibitors. We believe PARP inhibition results in an increase in unresolved spontaneous DNA single-strand breaks (SSBs), which collapse replication forks and trigger homologous recombination repair (HRR). We show that ATM is activated following inhibition of PARP. Furthermore, PARP inhibitor-induced HRR is abolished in ATM, but not DNA-PK, inhibited cells. ATM and DNA-PK inhibition together give the same sensitivity to PARP inhibitors as ATM alone, indicating that ATM functions in the same pathways as DNA-PK for survival at collapsed forks, likely in non-homologous end joining (NHEJ). Altogether, we suggest that ATM is activated by PARP inhibitor-induced collapsed replication forks and may function upstream of HRR in the repair of certain types of double-strand breaks (DSBs).
Figures
Figure 1
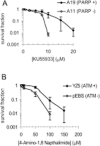
Absence of PARP-1 leads to increased sensitivity to ATM inhibition and vice versa. (A) Survival fraction of A19 (WT) and A11 (PARP-1−/−) MEFs following treatment for 10 days with increasing doses of the ATM inhibitor KU55933. (B) Survival fraction of AT221JE-T/pEBS (pEBS-ATM defective) and pEBS-YZ5 (YZ5—corrected for the AT phenotype by wild-type ATM cDNA) following treatment for 12 days with increasing doses of the PARP inhibitor 4-amino-1,8-napthalamide. The means (symbol) and standard deviations (error bar) from at least three experiments are depicted.
Figure 2
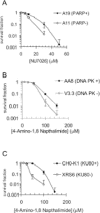
Absence of PARP-1 leads to increased sensitivity to DNA-PK inhibition and vice versa. (A) Survival fraction of A19 (WT) and A11 (PARP-1−/−) MEFs following treatment for 10 days with increasing doses of the DNA-PK inhibitor NU7026. (B) Survival fraction of AA8 (WT) and V3-3 (DNA-PKcs deficient) following treatment for 10 days with increasing doses of the PARP inhibitor 4-amino-1,8-napthalamide. (C) Survival fraction of CHO-K1 (WT) and XRS6 (Ku80 deficient) following treatment for 10 days with increasing doses of the PARP inhibitor 4-amino-1,8-napthalamide. The means (symbol) and standard deviations (error bar) from at least three experiments are depicted.
Figure 3
Inhibition of ATM and DNA-PK increases sensitivity to PARP inhibition compared to DNA-PK inhibition alone but not compared to ATM inhibition alone. Survival fraction of AA8 (WT) cells following treatment for 10 days with/without 10 µM KU55933 (ATM inhibitor), 10 µM NU7026 (DNA-PK inhibitor) or both in combination with increasing doses of the PARP inhibitor 4-amino-1,8-napthalamide. The means (symbol) and standard deviations (error bar) from at least three experiments are depicted.
Figure 4
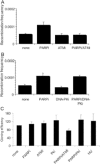
PARP inhibition activates ATM. (A) Western blot for ATM phospho serine 1981 and ATM control following PARP inhibition for the times indicated. (B) Western blot for CHK1 phospho serine 345, CHK2 phospho threonine 68 and total CHK2 and actin controls following PARP inhibition or 0.5 mM HU treatment for 24 h.
Figure 5
ATM inhibition prevents PARP inhibitor-induced HR. (A and B) Recombination frequency in hprt gene following treatment for 24 h with/without 10 µM KU55933 (ATM inhibitor), 10 µM NU7026 (DNA-PK inhibitor), 100 µM 4-amino-1,8-napthalamide (PARP inhibitor), 0.5 mM HU or combinations of the above. (C) Cloning efficiencies (% of control) of the same cells. The means (symbol) and standard deviations (error bar) from at least three experiments are depicted.
Figure 6
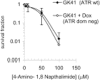
A kinase dead dominant negative ATR does not alter sensitivity to PARP inhibition. Survival fraction of GK41 cells following treatment for 10 days with increasing doses of the PARP inhibitor 4-amino-1,8-napthalamide in the presence or absence of 1.5 µg/ml doxycycline to induce expression of kinase dead dominant negative ATR. The means (symbol) and standard deviations (error bar) from at least three experiments are depicted.
Figure 7
Model for ATM activation following PARP inhibition. PARP inhibition results in more collapsed replication forks, probably because of an inability to efficiently repair endogenous SSBs (20). HR is the most important pathway for repair of collapsed replication forks in mammalian cells (32,33) and loss of this pathway results in lethality following PARP inhibition (20). Our data suggests that the HRR pathway involves an ATM signal, which would explain the increased sensitivity to PARP inhibitors in AT cells. In addition our data also imply that a second pathway involving DNA-PK and ATM is required in survival following PARP inhibition, this is most likely to be NHEJ.
Similar articles
- Role of autophagy in chemoresistance: regulation of the ATM-mediated DNA-damage signaling pathway through activation of DNA-PKcs and PARP-1.
Yoon JH, Ahn SG, Lee BH, Jung SH, Oh SH. Yoon JH, et al. Biochem Pharmacol. 2012 Mar 15;83(6):747-57. doi: 10.1016/j.bcp.2011.12.029. Epub 2011 Dec 29. Biochem Pharmacol. 2012. PMID: 22226932 - Requirement for NBS1 in the S phase checkpoint response to DNA methylation combined with PARP inhibition.
Horton JK, Stefanick DF, Zeng JY, Carrozza MJ, Wilson SH. Horton JK, et al. DNA Repair (Amst). 2011 Feb 7;10(2):225-34. doi: 10.1016/j.dnarep.2010.11.003. Epub 2010 Dec 3. DNA Repair (Amst). 2011. PMID: 21130714 Free PMC article. - Interaction between ATM and PARP-1 in response to DNA damage and sensitization of ATM deficient cells through PARP inhibition.
Aguilar-Quesada R, Muñoz-Gámez JA, Martín-Oliva D, Peralta A, Valenzuela MT, Matínez-Romero R, Quiles-Pérez R, Menissier-de Murcia J, de Murcia G, Ruiz de Almodóvar M, Oliver FJ. Aguilar-Quesada R, et al. BMC Mol Biol. 2007 Apr 25;8:29. doi: 10.1186/1471-2199-8-29. BMC Mol Biol. 2007. PMID: 17459151 Free PMC article. - The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer.
Smith J, Tho LM, Xu N, Gillespie DA. Smith J, et al. Adv Cancer Res. 2010;108:73-112. doi: 10.1016/B978-0-12-380888-2.00003-0. Adv Cancer Res. 2010. PMID: 21034966 Review. - DNA repair factors and telomere-chromosome integrity in mammalian cells.
Hande MP. Hande MP. Cytogenet Genome Res. 2004;104(1-4):116-22. doi: 10.1159/000077475. Cytogenet Genome Res. 2004. PMID: 15162024 Review.
Cited by
- Radiation-induced double-strand breaks require ATM but not Artemis for homologous recombination during S-phase.
Köcher S, Rieckmann T, Rohaly G, Mansour WY, Dikomey E, Dornreiter I, Dahm-Daphi J. Köcher S, et al. Nucleic Acids Res. 2012 Sep 1;40(17):8336-47. doi: 10.1093/nar/gks604. Epub 2012 Jun 22. Nucleic Acids Res. 2012. PMID: 22730303 Free PMC article. - 53BP1 depletion causes PARP inhibitor resistance in ATM-deficient breast cancer cells.
Hong R, Ma F, Zhang W, Yu X, Li Q, Luo Y, Zhu C, Jiang W, Xu B. Hong R, et al. BMC Cancer. 2016 Sep 9;16(1):725. doi: 10.1186/s12885-016-2754-7. BMC Cancer. 2016. PMID: 27613518 Free PMC article. - Cetuximab augments cytotoxicity with poly (adp-ribose) polymerase inhibition in head and neck cancer.
Nowsheen S, Bonner JA, Lobuglio AF, Trummell H, Whitley AC, Dobelbower MC, Yang ES. Nowsheen S, et al. PLoS One. 2011;6(8):e24148. doi: 10.1371/journal.pone.0024148. Epub 2011 Aug 30. PLoS One. 2011. PMID: 21912620 Free PMC article. - Inhibitors of poly ADP-ribose polymerase (PARP) induce apoptosis of myeloid leukemic cells: potential for therapy of myeloid leukemia and myelodysplastic syndromes.
Gaymes TJ, Shall S, MacPherson LJ, Twine NA, Lea NC, Farzaneh F, Mufti GJ. Gaymes TJ, et al. Haematologica. 2009 May;94(5):638-46. doi: 10.3324/haematol.2008.001933. Haematologica. 2009. PMID: 19407318 Free PMC article. - Pharmacological ascorbate induces 'BRCAness' and enhances the effects of Poly(ADP-Ribose) polymerase inhibitors against BRCA1/2 wild-type ovarian cancer.
Ma Y, Chen P, Drisko JA, Khabele D, Godwin AK, Chen Q. Ma Y, et al. Oncol Lett. 2020 Apr;19(4):2629-2638. doi: 10.3892/ol.2020.11364. Epub 2020 Jan 31. Oncol Lett. 2020. PMID: 32218813 Free PMC article.
References
- Lindahl T., Satoh M.S., Poirier G.G., Klungland A. Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem. Sci. 1995;20:405–411. - PubMed
- de Murcia J.M., Niedergang C., Trucco C., Ricoul M., Dutrillaux B., Mark M., Oliver F.J., Masson M., Dierich A., LeMeur M., Walztinger C., Chambon P., de Murcia G. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl Acad. Sci. USA. 1997;94:7303–7307. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous