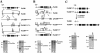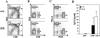Proteasome activator PA200 is required for normal spermatogenesis - PubMed (original) (raw)
Proteasome activator PA200 is required for normal spermatogenesis
Bernard Khor et al. Mol Cell Biol. 2006 Apr.
Abstract
The PA200 proteasome activator is a broadly expressed nuclear protein. Although how PA200 normally functions is not fully understood, it has been suggested to be involved in the repair of DNA double-strand breaks (DSBs). The PA200 gene (Psme4) is composed of 45 coding exons spanning 108 kb on mouse chromosome 11. We generated a PA200 null allele (PA200(Delta)) through Cre-loxP-mediated interchromosomal recombination after targeting loxP sites at either end of the locus. PA200(Delta/Delta) mice are viable and have no obvious developmental abnormalities. Both lymphocyte development and immunoglobulin class switching, which rely on the generation and repair of DNA DSBs, are unperturbed in PA200(Delta/Delta) mice. Additionally, PA200(Delta/Delta) embryonic stem cells do not exhibit increased sensitivity to either ionizing radiation or bleomycin. Thus, PA200 is not essential for the repair of DNA DSBs generated in these settings. Notably, loss of PA200 led to a marked reduction in male, but not female, fertility. This was due to defects in spermatogenesis observed in meiotic spermatocytes and during the maturation of postmeiotic haploid spermatids. Thus, PA200 serves an important nonredundant function during spermatogenesis, suggesting that the efficient generation of male gametes has distinct protein metabolic requirements.
Figures
FIG. 1.
Generation of the PA200Δ allele. A and B. Schematic of the murine PA200 locus, drawn approximately to scale. The targeting constructs pPA2005′LoxP.Neo and pPA2003′LoxP.Neo are shown, with relevant features indicated, including the neomycin resistance gene (N), flanking loxP sites (open ovals), and thymidine kinase gene (TK). Relevant endogenous and introduced (*) BamHI (B and B*) and HindIII (H and H*) sites are shown. Also shown are schematics of the targeted alleles before (PA2005′LoxP.Neo and PA2003′LoxP.Neo) and after (PA2005′LoxP and PA2003′LoxP) Cre deletion of the neomycin resistance gene. The relative positions of the P1, P2, and P3 probes are shown along with Southern blot analyses of either BamHI- (B) or HindIII- (H) digested ES cell genomic DNA. The bands representing the different targetings are indicated with arrows. The BamHI P3-hybridizing bands from the PA200+ and PA2003′LoxP.Neo alleles are of similar size and are indicated with a single arrow. DNA molecular mass markers (in kb) are also indicated. C. Schematic of Cre-mediated recombination between the PA2005′LoxP and PA2003′LoxP alleles, generating the PA200Δ and PA200RP alleles through a chromosomal translocation. Southern blot analyses of EcoRV (V)-digested ES cell genomic DNA probed with P1 and P3 are shown. The bands generated by the PA200Δ (Δ) and PA200RP (RP) alleles in the PA200-227 (227) ES cell line are indicated.
FIG. 2.
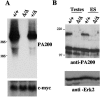
PA200 mRNA and protein are not detected in PA200Δ/Δ cells. A. Northern blot analyses of mRNA from PA200+/+, PA200+/Δ, and PA200Δ/Δ ES cells using a PA200 cDNA probe and a c-myc probe as an RNA loading control. B. Western blot analyses of total cell lysates from PA200+/+ and PA200Δ/Δ ES cells and testes using an anti-PA200 antibody and an anti-Erk2 antibody as a protein loading control.
FIG. 3.
Lymphocyte development and immunoglobulin class switch recombination in PA200Δ/Δ mice. A, B, and C. Flow cytometric analyses of thymocytes (A) and splenocytes (B and C) from PA200+/+ and PA200Δ/Δ mice using either anti-CD4 and anti-CD8 (A and B) or anti-Thy1.2 and anti-B220 (C) antibodies. Gates and percentages are shown. D. Concentration of IgG1 in the supernatant of purified PA200+/+ (filled bars) and PA200Δ/Δ (open bars) B cells stimulated with either LPS or LPS plus IL-4. IgG1 concentrations were determined by ELISA. The mean concentrations and standard deviations from two experiments are shown.
FIG. 4.
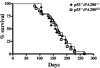
Kaplan-Meier survival curves for PA200+/+ p53−/− (filled diamonds; n = 21) and PA200Δ/Δ p53−/− (open squares; n = 14) mice.
FIG. 5.
Histology of PA200Δ/Δ testes and epidydimides. A. Low magnification of testes (plates A and B; bar, 100 μm) and epididymis (plates C and D; bar, 50 μm) from 5-month-old PA200+/+ (wild type [WT]; plates A and C) and PA200Δ/Δ (plates B and E) mice. Areas showing vacuolation, multinucleated giant germ cells, and hypospermatocytogenesis are indicated (plate B, circles). Immature germ cells in the PA200Δ/Δ caput epididymis are indicated (plate D, arrows). B. Seminiferous tubules from PA200+/+ (WT; plates A and B) and PA200Δ/Δ (Δ/Δ; plates C to H) mice. Bar, 50 μm. Histologic stages of development are indicated with roman numerals. Stages of spermatid maturation are indicated with Arabic numbers. Also indicated are pachytene (P) and leptotene (L) spermatocytes, Sertoli cells (Sc), B-type spermatogonia (B), areas of vacuolation (V), and residual bodies (Rb). The arrows indicate residual bodies (plates C and E), apoptotic and necrotic pachytene spermatocytes (plates D and F), and spermatid phagocytosis (plate G). The asterisk indicates abnormal spermatids (plate C) and abnormal pachetyne spermatocytes (plate E). Abnormal spermatid heads in the seminiferous epithelium (plate H, circle) are shown.
FIG. 6.
Increased apoptotic cells in PA200Δ/Δ testes. A. Mean ± standard deviation number of TUNEL-positive cells per seminiferous tubule section in PA200+/+ (open bar; n = 2) and PA200Δ/Δ (filled bar; n = 4) mice. A minimum of 125 seminiferous tubule sections was counted from each mouse. B. TUNEL staining of testes sections from PA200+/+ and PA200Δ/Δ mice. Shown are the sections containing the seminiferous tubule, with the greatest number of TUNEL-positive cells from +/+ and Δ/Δ mice.
Similar articles
- The PSMA8 subunit of the spermatoproteasome is essential for proper meiotic exit and mouse fertility.
Gómez-H L, Felipe-Medina N, Condezo YB, Garcia-Valiente R, Ramos I, Suja JA, Barbero JL, Roig I, Sánchez-Martín M, de Rooij DG, Llano E, Pendas AM. Gómez-H L, et al. PLoS Genet. 2019 Aug 22;15(8):e1008316. doi: 10.1371/journal.pgen.1008316. eCollection 2019 Aug. PLoS Genet. 2019. PMID: 31437213 Free PMC article. - Proteasome subunit α4s is essential for formation of spermatoproteasomes and histone degradation during meiotic DNA repair in spermatocytes.
Zhang ZH, Jiang TX, Chen LB, Zhou W, Liu Y, Gao F, Qiu XB. Zhang ZH, et al. J Biol Chem. 2021 Jan-Jun;296:100130. doi: 10.1074/jbc.RA120.016485. Epub 2020 Dec 4. J Biol Chem. 2021. PMID: 33262216 Free PMC article. - Ku70 and non-homologous end joining protect testicular cells from DNA damage.
Ahmed EA, Sfeir A, Takai H, Scherthan H. Ahmed EA, et al. J Cell Sci. 2013 Jul 15;126(Pt 14):3095-104. doi: 10.1242/jcs.122788. J Cell Sci. 2013. PMID: 23857907 Free PMC article. - In vitro spermatogenesis: Why meiotic checkpoints matter.
Lei Q, van Pelt AMM, Hamer G. Lei Q, et al. Curr Top Dev Biol. 2023;151:345-369. doi: 10.1016/bs.ctdb.2022.04.009. Epub 2022 Jun 9. Curr Top Dev Biol. 2023. PMID: 36681476 Review. - PA200-Mediated Proteasomal Protein Degradation and Regulation of Cellular Senescence.
Wen P, Sun Y, Jiang TX, Qiu XB. Wen P, et al. Int J Mol Sci. 2024 May 22;25(11):5637. doi: 10.3390/ijms25115637. Int J Mol Sci. 2024. PMID: 38891826 Free PMC article. Review.
Cited by
- Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis.
Qian MX, Pang Y, Liu CH, Haratake K, Du BY, Ji DY, Wang GF, Zhu QQ, Song W, Yu Y, Zhang XX, Huang HT, Miao S, Chen LB, Zhang ZH, Liang YN, Liu S, Cha H, Yang D, Zhai Y, Komatsu T, Tsuruta F, Li H, Cao C, Li W, Li GH, Cheng Y, Chiba T, Wang L, Goldberg AL, Shen Y, Qiu XB. Qian MX, et al. Cell. 2013 May 23;153(5):1012-24. doi: 10.1016/j.cell.2013.04.032. Cell. 2013. PMID: 23706739 Free PMC article. - Queen conch (Strombus gigas) testis regresses during the reproductive season at nearshore sites in the Florida Keys.
Spade DJ, Griffitt RJ, Liu L, Brown-Peterson NJ, Kroll KJ, Feswick A, Glazer RA, Barber DS, Denslow ND. Spade DJ, et al. PLoS One. 2010 Sep 15;5(9):e12737. doi: 10.1371/journal.pone.0012737. PLoS One. 2010. PMID: 20856805 Free PMC article. - Essential Role of Histone Replacement and Modifications in Male Fertility.
Wang T, Gao H, Li W, Liu C. Wang T, et al. Front Genet. 2019 Oct 8;10:962. doi: 10.3389/fgene.2019.00962. eCollection 2019. Front Genet. 2019. PMID: 31649732 Free PMC article. Review. - The PSMA8 subunit of the spermatoproteasome is essential for proper meiotic exit and mouse fertility.
Gómez-H L, Felipe-Medina N, Condezo YB, Garcia-Valiente R, Ramos I, Suja JA, Barbero JL, Roig I, Sánchez-Martín M, de Rooij DG, Llano E, Pendas AM. Gómez-H L, et al. PLoS Genet. 2019 Aug 22;15(8):e1008316. doi: 10.1371/journal.pgen.1008316. eCollection 2019 Aug. PLoS Genet. 2019. PMID: 31437213 Free PMC article. - Structure of the Blm10-20 S proteasome complex by cryo-electron microscopy. Insights into the mechanism of activation of mature yeast proteasomes.
Iwanczyk J, Sadre-Bazzaz K, Ferrell K, Kondrashkina E, Formosa T, Hill CP, Ortega J. Iwanczyk J, et al. J Mol Biol. 2006 Oct 27;363(3):648-59. doi: 10.1016/j.jmb.2006.08.010. Epub 2006 Aug 9. J Mol Biol. 2006. PMID: 16952374 Free PMC article.
References
- Ahn, J. Y., N. Tanahashi, K. Akiyama, H. Hisamatsu, C. Noda, K. Tanaka, C. H. Chung, N. Shibmara, P. J. Willy, J. D. Mott, et al. 1995. Primary structures of two homologous subunits of PA28, a gamma-interferon-inducible protein activator of the 20S proteasome. FEBS Lett. 366:37-42. - PubMed
- Aifantis, I., C. Borowski, F. Gounari, H. D. Lacorazza, J. Nikolich-Zugich, and H. von Boehmer. 2002. A critical role for the cytoplasmic tail of pTα in T lymphocyte development. Nat. Immunol. 3:483-488. - PubMed
- Alfonso, P. J., and W. S. Kistler. 1993. Immunohistochemical localization of spermatid nuclear transition protein 2 in the testes of rats and mice. Biol. Reprod. 48:522-529. - PubMed
- Baarends, W. M., J. W. Hoogerbrugge, H. P. Roest, M. Ooms, J. Vreeburg, J. H. Hoeijmakers, and J. A. Grootegoed. 1999. Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Dev. Biol. 207:322-333. - PubMed
- Barton, L. F., H. A. Runnels, T. D. Schell, Y. Cho, R. Gibbons, S. S. Tevethia, G. S. Deepe, Jr., and J. J. Monaco. 2004. Immune defects in 28-kDa proteasome activator gamma-deficient mice. J. Immunol. 172:3948-3954. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI 47829/AI/NIAID NIH HHS/United States
- P30 AR048335/AR/NIAMS NIH HHS/United States
- RR 012466/RR/NCRR NIH HHS/United States
- P30 AR 48335/AR/NIAMS NIH HHS/United States
- R01 AI047829/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases