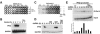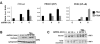Positive and negative regulation of the innate antiviral response and beta interferon gene expression by deacetylation - PubMed (original) (raw)
Positive and negative regulation of the innate antiviral response and beta interferon gene expression by deacetylation
Inna Nusinzon et al. Mol Cell Biol. 2006 Apr.
Abstract
Beta interferon (IFN-beta) gene expression in response to virus infection relies on the dynamic assembly of a multiprotein enhanceosome complex that is initiated by the activation of two inducible transcription factors, interferon regulatory factor 3 (IRF3) and NF-kappaB. Virus or double-stranded RNA-induced activation of IFN-beta gene expression is prevented by the addition of protein deacetylase inhibitors. The isolated IRF-responsive positive regulatory domain was found to require deacetylation for its activity, but IRF3 protein activation leading to its nuclear translocation and DNA binding was not impaired by deacetylase inhibition. In contrast, NF-kappaB activity was not affected by deacetylase inhibitors. RNA interference indicated that several deacetylase enzymes, including histone deacetylase 1 (HDAC1), HDAC8, and HDAC6, influence IFN-beta gene expression with opposing effects. While HDAC1 and HDAC8 repress IFN-beta expression, HDAC6 acts as a coactivator essential for enhancer activity. Virus replication is enhanced in HDAC6-depleted cells, demonstrating HDAC6 is an essential component of innate antiviral immunity.
Figures
FIG. 1.
Deacetylase activity is required for innate antiviral defense. A. Duplicate wells of 2fTGH cells were infected with dilutions of Sendai virus to achieve MOIs of 62.5, 12.5, 2.5, 0.5, 0.1, and 0 PFU/cell. Simultaneously with inoculation, cells were treated with TSA at the indicated concentrations. Cells were stained and analyzed 24 h postinfection. B. 2fTGH cells were infected with Sendai virus (MOI, 10 PFU/cell) for the indicated times in the presence or absence of TSA. RT-PCR was carried out using primers for IFN-β or the control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH). C. 2fTGH cells were pretreated with dsRNA for 8 h with or without TSA and then infected with VSV at MOIs of 50, 5, 0.5, 0.05, 0.005, and 0 PFU/cell. Cells were stained and analyzed 18 h postinfection. D. 2fTGH cells were treated with dsRNA, TSA, and cycloheximide (CHX) as indicated, and RT-PCR for IFN-β and GAPDH was performed. E. (Top) HeLa cells were treated with dsRNA and TSA as indicated, and ChIP assays were performed using antiserum specific for RNA Pol II. Coprecipitated DNA was amplified by PCR using primers specific for the IFN-β promoter. (Bottom) Phosphorimaging was used for quantitative analysis, and values were normalized to the input.
FIG. 2.
Deacetylase activity is required for IFN-β enhanceosome activity. A. Luciferase assays were performed in 2fTGH cells using the indicated reporter genes, and cells were treated with dsRNA and TSA for the indicated times. B. 2fTGH cells were treated with dsRNA and TSA as indicated, and nuclear and cytoplasmic fractions were separated followed by SDS-PAGE and Western blotting with antibodies for IRF3. C. 2fTGH cells were treated with dsRNA and TSA as indicated, nuclei were purified, and nuclear lysates were incubated with a biotinylated double-stranded oligonucleotide containing IRF3 binding sites. IRF3 was eluted from DNA and visualized by SDS-PAGE and Western blotting. Asterisks indicate the IRF3 band. Bound, IRF3 that was bound to oligonucleotide; total, total nuclear IRF3; c, control containing no oligonucleotide.
FIG. 3.
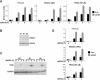
HDAC1 and HDAC8 inhibit IFN-β expression. A. Luciferase assays with the indicated reporters were performed in the presence of siRNA for HDAC1 (siHD1) or scrambled control (Con). At 48 h after transfection with siRNA and reporter, cells were treated with dsRNA for the indicated times. B. Two clones stably expressing RNA interference vectors specific for HDAC1 were analyzed by Western blotting. wt, wild-type cells. C. The stable clones were treated with dsRNA as indicated, and RT-PCR was performed for IFN-β or glyceraldehyde-3-phosphate dehydrogenase (GAPDH). WT, wild-type cells. D. Similar to the experiment in panel A, except siRNA specific for HDAC8 was used.
FIG. 4.
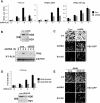
HDAC6 is required for IFN-β expression and innate antiviral immunity. A. Luciferase assays with the indicated reporters were performed in the presence of siRNA for HDAC6 (siHD6) or scrambled control (Con). At 48 h after transfection with siRNA and reporter, cells were treated with dsRNA for the indicated times. B. (Top) Western blot for HDAC6 in 2fTGH cells transfected with HDAC6-specific siRNA. (Bottom) 2fTGH cells transfected with siRNA were treated with dsRNA, and RT-PCR was performed for IFN-β and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). WB, Western blot. C. 2fTGH cells were transfected with siRNA and 48 h later were treated with dsRNA (8 h) or Sendai virus (5 h) or left untreated (unt) and then infected with VSV-GFP for 18 h. D. (Top) Luciferase assay similar to the experiment shown in panel A, but with A549 cells (control) or an HDAC6-silenced A549 stable cell line (HD6 KD). (Bottom) Western blot for HDAC6 in the A549 cells above. E. Similar to the experiment in panel C, but with A549 cells (Con) or a HDAC6-silenced A549 stable cell line (HD6 KD).
FIG. 5.
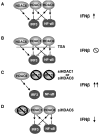
HDACs are activators and repressors of IFN-β. A. HDAC6 activates IRF3, while HDAC1 and HDAC8 repress both IRF3 and NF-κB. Together, these HDACs strike a balance for optimal activation of IFN-β. B. TSA inhibits all HDAC activity, thereby blocking IFN-β activation. C. siRNA specific for either HDAC1 or HDAC8 relieves the inhibitory actions of these proteins, resulting in a heightened transcriptional response. D. siRNA specific for HDAC6 removes the activating effect of this deacetylase, causing lower IFN-β expression.
Similar articles
- Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1.
Nusinzon I, Horvath CM. Nusinzon I, et al. Proc Natl Acad Sci U S A. 2003 Dec 9;100(25):14742-7. doi: 10.1073/pnas.2433987100. Epub 2003 Nov 25. Proc Natl Acad Sci U S A. 2003. PMID: 14645718 Free PMC article. - PKC alpha regulates Sendai virus-mediated interferon induction through HDAC6 and β-catenin.
Zhu J, Coyne CB, Sarkar SN. Zhu J, et al. EMBO J. 2011 Sep 27;30(23):4838-49. doi: 10.1038/emboj.2011.351. EMBO J. 2011. PMID: 21952047 Free PMC article. - Histone deacetylase 1/mSin3A disrupts gamma interferon-induced CIITA function and major histocompatibility complex class II enhanceosome formation.
Zika E, Greer SF, Zhu XS, Ting JP. Zika E, et al. Mol Cell Biol. 2003 May;23(9):3091-102. doi: 10.1128/MCB.23.9.3091-3102.2003. Mol Cell Biol. 2003. PMID: 12697811 Free PMC article. - The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression.
Ashburner BP, Westerheide SD, Baldwin AS Jr. Ashburner BP, et al. Mol Cell Biol. 2001 Oct;21(20):7065-77. doi: 10.1128/MCB.21.20.7065-7077.2001. Mol Cell Biol. 2001. PMID: 11564889 Free PMC article. - Regulation of the MIR155 host gene in physiological and pathological processes.
Elton TS, Selemon H, Elton SM, Parinandi NL. Elton TS, et al. Gene. 2013 Dec 10;532(1):1-12. doi: 10.1016/j.gene.2012.12.009. Epub 2012 Dec 14. Gene. 2013. PMID: 23246696 Review.
Cited by
- Acetylation licenses Th1 cell polarization to constrain Listeria monocytogenes infection.
Zhang YS, Xin DE, Wang Z, Peng W, Zeng Y, Liang J, Xu M, Chen N, Zhang J, Yue J, Cao M, Zhang C, Wang Y, Chang Z, Lu XM, Chang L, Chinn YE. Zhang YS, et al. Cell Death Differ. 2022 Nov;29(11):2303-2315. doi: 10.1038/s41418-022-01017-9. Epub 2022 May 25. Cell Death Differ. 2022. PMID: 35614130 Free PMC article. - Histone deacetylase 11 regulates oligodendrocyte-specific gene expression and cell development in OL-1 oligodendroglia cells.
Liu H, Hu Q, D'ercole AJ, Ye P. Liu H, et al. Glia. 2009 Jan 1;57(1):1-12. doi: 10.1002/glia.20729. Glia. 2009. PMID: 18627006 Free PMC article. - Pharmacologic and chemical adjuvants in tumor virotherapy.
Alvarez-Breckenridge C, Kaur B, Chiocca EA. Alvarez-Breckenridge C, et al. Chem Rev. 2009 Jul;109(7):3125-40. doi: 10.1021/cr900048k. Chem Rev. 2009. PMID: 19462957 Free PMC article. Review. No abstract available. - Combining Oncolytic Viruses and Small Molecule Therapeutics: Mutual Benefits.
Spiesschaert B, Angerer K, Park J, Wollmann G. Spiesschaert B, et al. Cancers (Basel). 2021 Jul 6;13(14):3386. doi: 10.3390/cancers13143386. Cancers (Basel). 2021. PMID: 34298601 Free PMC article. Review. - Inhibition of viral pathogenesis and promotion of the septic shock response to bacterial infection by IRF-3 are regulated by the acetylation and phosphorylation of its coactivators.
Chattopadhyay S, Fensterl V, Zhang Y, Veleeparambil M, Wetzel JL, Sen GC. Chattopadhyay S, et al. mBio. 2013 Mar 26;4(2):e00636-12. doi: 10.1128/mBio.00636-12. mBio. 2013. PMID: 23532979 Free PMC article.
References
- Agalioti, T., G. Chen, and D. Thanos. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111:381-392. - PubMed
- Bhattacharya, S., R. Eckner, S. Grossman, E. Oldread, Z. Arany, A. D'Andrea, and D. M. Livingston. 1996. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-alpha. Nature 383:344-347. - PubMed
- Chambers, A. E., S. Banerjee, T. Chaplin, J. Dunne, S. Debernardi, S. P. Joel, and B. D. Young. 2003. Histone acetylation-mediated regulation of genes in leukaemic cells. Eur. J. Cancer 39:1165-1175. - PubMed
- Cohen, T., and T. P. Yao. 2004. AcK-knowledge reversible acetylation. Sci. STKE 2004:pe42. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous